 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00013451-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 430aa MW: 46717.7 Da PI: 6.4603 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 31.9 | 2.8e-10 | 235 | 292 | 5 | 62 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklkse 62
kr++r NR +A rs +RK +i eLe+kv++L++e ++L +l l+ l++e
WALNUT_00013451-RA 235 KRAKRIWANRQSAARSKERKMRYISELERKVQTLQTEATSLSAQLTLLQRDTNGLTAE 292
9********************************************9999887777766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.7E-15 | 231 | 295 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.818 | 233 | 296 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 5.1E-11 | 235 | 289 | No hit | No description |
| SuperFamily | SSF57959 | 3.94E-11 | 235 | 289 | No hit | No description |
| Pfam | PF00170 | 9.3E-9 | 235 | 287 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14703 | 1.45E-24 | 236 | 285 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 430 aa Download sequence Send to blast |
MDKEKTPGHG GGLPPPSGRY SGFSPTGSSF NVKPEPSSAS PSYPPLAPGS SSDSAHFGHG 60 MSADSSRFSH DISRMPDTPR RSLGHRRAHS EILTLPDDIS FDSDLGVVGG ADGPSFSDET 120 EEDLLSMYLD MDKFNSSSAT SSFQVGEPSS AAAAPSPALA AMPTPASGAV TSSENFAIGS 180 NERPRVRHQH SQSMDGSTTI KPEMLVSGSE DMSAADSKKA MSAAKLAELA LIDPKRAKRI 240 WANRQSAARS KERKMRYISE LERKVQTLQT EATSLSAQLT LLQRDTNGLT AENGELKLRL 300 QTMEQQVHLQ DALNDALKEE IQHLKVLTGQ AIPNGGPMMN FASFGAGQQF YPNNHAMHTL 360 LTTQQFQQLQ IHSQKQQQQF QQHQLHQLQQ QQLQQQQQEQ QQQQTGDLKI RGSLSSPSQK 420 ENATDVDSAV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
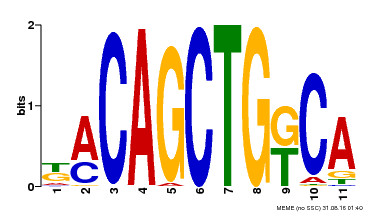
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018839959.1 | 0.0 | PREDICTED: probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-159 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A2I4G7U1 | 0.0 | A0A2I4G7U1_JUGRE; probable transcription factor PosF21 | ||||
| STRING | XP_008222595.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5517 | 32 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-144 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



