 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00016004-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 589aa MW: 65706.9 Da PI: 7.2108 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 33.6 | 8.7e-11 | 280 | 315 | 4 | 39 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLe 39
k rr+++NReAAr+sR+RKka++++Le +L+
WALNUT_00016004-RA 280 AKTLRRLAQNREAARKSRLRKKAYVQQLESSRMKLT 315
6889*************************8766665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 3.5E-8 | 272 | 323 | No hit | No description |
| SMART | SM00338 | 2.0E-6 | 277 | 354 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.059 | 279 | 323 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 5.0E-8 | 280 | 318 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.13E-6 | 281 | 323 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 284 | 299 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 8.4E-30 | 363 | 436 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 589 aa Download sequence Send to blast |
MMQESQAFNK KQDDNSTQSA MNLLQLRTVE GQQKFLLMER NLNVAWSSFL DENENGLRWS 60 CMASHRVGPV GETGLSDSGP SNPHVPYSVF HGSNVPTSFI NQEGSAFDFR ELEEAIVLQG 120 VKIRNDEAKA PLFTGRPAAT LEMFPSWPMR FQTTPRVVVA SNFSPSLTNL LIFWFVLHQG 180 SSKSGGESTD SGSAVNTFSS KAEVQLESES PISNKASSSD HQAFDHQKHL QLQQQQIQQE 240 MANDRSRAGP SQNQSDVKAP QEKRKGSGGS SSTSEKQLDA KTLRRLAQNR EAARKSRLRK 300 KAYVQQLESS RMKLTHIEQD LQRARAQGLF LGGCGGAGGN LSSGAAIFDM EYARWLEDDH 360 RLMAELRTGL HAHLSDGDLR LIVEGYISHY DEIFRLKGVA AKSDVFHVIT GLWMTPAERC 420 FLWIGGFRPS DLIKMLIGQL EPLTEQQLVG IYRLQQSTQQ AEEALSQGQE QLQQSLIETI 480 AAAGPVIDGM QQMVVALGKL SNLEGFVRQA DNLRQQTLHQ LRHVLTIRQA ARCFLVIGEY 540 YGRLRALSSL WASRPRESLM SEDHNSCQTT ATDHLQIVHQ SAHNHFSTF |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Together with TGA10, basic leucine-zipper transcription factor required for anther development, probably via the activation of SPL expression in anthers and via the regulation of genes with functions in early and middle tapetal development (PubMed:20805327). Required for signaling responses to pathogen-associated molecular patterns (PAMPs) such as flg22 that involves chloroplastic reactive oxygen species (ROS) production and subsequent expression of H(2)O(2)-responsive genes (PubMed:27717447). {ECO:0000269|PubMed:20805327, ECO:0000269|PubMed:27717447}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
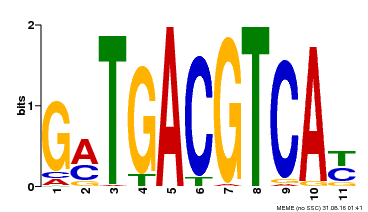
| Motif ID | Method | Source | Motif file |
| MP00131 | DAP | Transfer from AT1G08320 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by flg22 in leaves. {ECO:0000269|PubMed:27717447}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018813854.1 | 0.0 | PREDICTED: transcription factor TGA2.2-like isoform X1 | ||||
| Swissprot | Q93XM6 | 0.0 | TGA9_ARATH; Transcription factor TGA9 | ||||
| TrEMBL | A0A2I4E378 | 0.0 | A0A2I4E378_JUGRE; transcription factor TGA2.2-like isoform X1 | ||||
| STRING | EOY33449 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4555 | 33 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08320.3 | 1e-178 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



