 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00018329-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 332aa MW: 36353.3 Da PI: 7.1929 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 108.6 | 3.1e-34 | 182 | 239 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s+fprsYYrCt+++C+vkk+vers +dp++v++tYeg+Hnh+
WALNUT_00018329-RA 182 EDGYRWRKYGQKAVKNSPFPRSYYRCTTQKCTVKKRVERSFQDPSTVITTYEGQHNHP 239
8********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.7E-35 | 166 | 241 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 6.67E-30 | 173 | 241 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.569 | 176 | 241 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.0E-40 | 181 | 240 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.2E-26 | 182 | 239 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
MSDESKDQLY FHEYLFQDNQ NLASGSSSTG SYNHKPPASI AAANSSAYSS QGFDPSNYMS 60 FTECLQTPMD YNSMASAFGL SPSSSEVFAS MESNPKPMEV GDLGGGGSDI ILVTPNSSIS 120 SSSTEVGAEE DSGKSSKDKQ PKETEVGGES SKKVSKAKKK GDKKQREPRF AFMTKSEVDH 180 LEDGYRWRKY GQKAVKNSPF PRSYYRCTTQ KCTVKKRVER SFQDPSTVIT TYEGQHNHPI 240 PATLRGNAAA MSPPSMFTPP IGGRPTFIPQ DFFAQMPQIM SSSTSQAGGG TAPGSINYSQ 300 INPNYPHHQQ YQLPDYGLLQ DIIPSMFLKQ EP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-26 | 174 | 242 | 10 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-26 | 174 | 242 | 10 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
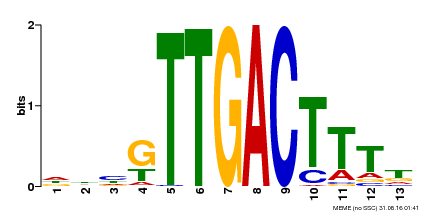
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00440 | DAP | Transfer from AT4G18170 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018841904.1 | 0.0 | PREDICTED: probable WRKY transcription factor 28 | ||||
| Swissprot | Q8VWJ2 | 2e-77 | WRK28_ARATH; WRKY transcription factor 28 | ||||
| TrEMBL | A0A2I4GDC9 | 0.0 | A0A2I4GDC9_JUGRE; probable WRKY transcription factor 28 | ||||
| STRING | cassava4.1_011936m | 1e-118 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2231 | 34 | 83 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18170.1 | 3e-66 | WRKY DNA-binding protein 28 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



