 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00020265-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 871aa MW: 96556.5 Da PI: 6.6488 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.6 | 2.1e-18 | 77 | 135 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++ +++ps +r++L +++ +++ +q+kvWFqNrR +ek+
WALNUT_00020265-RA 77 KYVRYTPEQVEALERLYHDCPKPSSIRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 135
5679*****************************************************97 PP
| |||||||
| 2 | START | 176.5 | 1.6e-55 | 221 | 429 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..ga 90
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++++++l+v+ ++ g+
WALNUT_00020265-RA 221 IAEETLAEFLSKATGTAVEWVQMPGMKPGPDSIGIVAISHGCTGVAARACGLVGLEPT-RVAEILKDRPSWFRDCRAVDVLNVLPTAngGT 310
7899******************************************************.8888888888****************9999** PP
EEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXX CS
START 91 lqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlp 177
++l +++l+a+++l+p Rdf+ +Ry+ l++g++v++++S+ ++q+ p+ +++vRae+lpSg+li+p+++ +s +++v+h+dl+ +++
WALNUT_00020265-RA 311 IELLYMQLYAPTTLAPaRDFWLLRYTSVLEDGSLVVCERSLKNTQNGPSmppVQHFVRAEMLPSGYLIRPCEGSGSIIHIVDHMDLEPWSV 401
***********************************************9988899************************************* PP
HHHHHHHHHHHHHHHHHHHHHHTXXXXX CS
START 178 hwllrslvksglaegaktwvatlqrqce 205
+++lr+l++s+++ ++kt++ +l+++++
WALNUT_00020265-RA 402 PEVLRPLYESSTVLAQKTTMTALRQLRQ 429
***********************99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.548 | 72 | 136 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-15 | 74 | 140 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.15E-16 | 76 | 140 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.82E-16 | 77 | 137 | No hit | No description |
| Pfam | PF00046 | 6.1E-16 | 78 | 135 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-18 | 79 | 135 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.47E-6 | 129 | 168 | No hit | No description |
| PROSITE profile | PS50848 | 25.02 | 211 | 426 | IPR002913 | START domain |
| CDD | cd08875 | 3.46E-78 | 215 | 431 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 2.2E-23 | 220 | 427 | IPR023393 | START-like domain |
| SMART | SM00234 | 3.7E-40 | 220 | 430 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.79E-37 | 220 | 431 | No hit | No description |
| Pfam | PF01852 | 5.4E-53 | 221 | 429 | IPR002913 | START domain |
| Pfam | PF08670 | 2.1E-52 | 728 | 870 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 871 aa Download sequence Send to blast |
MNNSESVAGT RSDNAELDLD TRLCRWIDFR FVSLLLLWSS SKCFELSLRK QSYLDFSWLF 60 WMAMSCKDGK LAMDNGKYVR YTPEQVEALE RLYHDCPKPS SIRRQQLIRE CPILSNIEPK 120 QIKVWFQNRR CREKQRKEAS RLQAVNRKLT AMNKLLMEEN DRLQKQVSHL VYENGYFRQH 180 TQNTTLATKD TSCESVVTSG QHHLTAQHPP RDASPAGLLS IAEETLAEFL SKATGTAVEW 240 VQMPGMKPGP DSIGIVAISH GCTGVAARAC GLVGLEPTRV AEILKDRPSW FRDCRAVDVL 300 NVLPTANGGT IELLYMQLYA PTTLAPARDF WLLRYTSVLE DGSLVVCERS LKNTQNGPSM 360 PPVQHFVRAE MLPSGYLIRP CEGSGSIIHI VDHMDLEPWS VPEVLRPLYE SSTVLAQKTT 420 MTALRQLRQI AHEVSQSNTT GWARRPAALR ALSQRLSRGF NEALNGFTDE GWSMMGNDGM 480 DDVTILVNSS PDKLMGLNLP FSNGFPAVSN AVLCAKASML LQNVPPAILL RFLREHRSEW 540 ADNNIDAYSA AAVKIGPCSL PGSRVGSFGG QVILPLAHTM EHEELCSGMD ENAVGTCAEL 600 IFAPIDASFA DDAPLLPSGF RIIPLDSGKE ASSPNRTLDL ASALEIGPAG NRTSTDYSAN 660 AGSVRSVMTI AFEFAYESHM QEHVASMARQ YVRSIISSVQ RVALALSPSH LSSHAGLRPP 720 LGTPEAQTLA RWICQSYRCY MGVELLKSGN EGSEANLKTL WHHSDAIMCC SMKALPVFTF 780 ANQAGLDMLE TTLVALQDIT LEKIFDDHGR KTLFSEFPQI MQQGFACLQG GICLSSMGRP 840 VSYERAVAWK VLNEEENAHC ICFMFVNWSF V |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
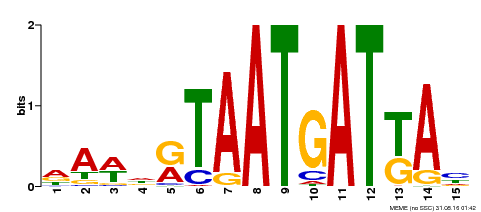
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018847857.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15 | ||||
| Refseq | XP_018847858.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A2I4GVE0 | 0.0 | A0A2I4GVE0_JUGRE; homeobox-leucine zipper protein ATHB-15 | ||||
| STRING | cassava4.1_001672m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1946 | 33 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



