 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00020277-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 366aa MW: 40930.4 Da PI: 8.3235 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 22.5 | 3e-07 | 210 | 232 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ Cgk F+r nL+ H+r H
WALNUT_00020277-RA 210 HYCQVCGKGFKRDANLRMHMRAH 232
68*******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 4.35E-6 | 208 | 236 | No hit | No description |
| SMART | SM00355 | 0.0034 | 210 | 232 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.653 | 210 | 237 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.6E-6 | 210 | 236 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 212 | 232 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 76 | 265 | 298 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 33 | 303 | 325 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 366 aa Download sequence Send to blast |
MLSATISCYP DVPQGLLQIS SMTEDIISSS LEVNSGTDSH PSSLLSNFSI LKDKVFQVES 60 LISILNSLNH SETELASSLA AVGMDTVIQE IIMTTSSMMF ICQQMALGFT SAGTSNHELQ 120 QQQQCRQNRF PQSNFGSNRD GIVIQEGEQG FYSGYMVEWN DENYNNSSNK DENRSVITGK 180 NKKAGDGQRL SLEYDVIQLE AADILAKYTH YCQVCGKGFK RDANLRMHMR AHGDEYKTIA 240 ALGNPMKNGS VLGNRKGLRE LPTKYSCPQE GCRWNQKHAK FQPLKSIVCV KNHYKRSHCP 300 KIYVCKRCNH KQFSVLSDLR THEKHCGDQK WQCSCGTTFS RKDKLMGHVA LFVGHTPASS 360 SFINLA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Together with STOP1, plays a critical role in tolerance to major stress factors in acid soils such as proton H(+) and aluminum ion Al(3+). Required for the expression of genes in response to acidic stress (e.g. ALMT1 and MATE), and Al-activated citrate exudation. {ECO:0000269|PubMed:23935008}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00522 | DAP | Transfer from AT5G22890 | Download |
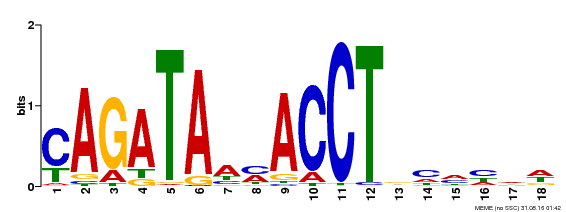
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018841940.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like | ||||
| Swissprot | Q0WT24 | 1e-104 | STOP2_ARATH; Protein SENSITIVE TO PROTON RHIZOTOXICITY 2 | ||||
| TrEMBL | A0A2I4GDI7 | 0.0 | A0A2I4GDI7_JUGRE; protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like | ||||
| STRING | EOY33679 | 1e-142 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8099 | 30 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22890.1 | 1e-101 | C2H2 family protein | ||||



