 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00020872-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 390aa MW: 44338.4 Da PI: 4.7853 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 112.3 | 3.3e-35 | 14 | 105 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkke 92
Fl+k+ye+++d+++++++sws+ ++sfvv+++ efa+ +Lpk+Fkh+nf+SF+RQLn+YgF+k++ e+ weF+++ F +g+ +
WALNUT_00020872-RA 14 FLSKTYEMVDDPSTDSVVSWSQCNKSFVVWNPPEFARVLLPKFFKHNNFSSFIRQLNTYGFRKIDPEQ---------WEFANEDFMRGQPH 95
9***************************************************************9998.........************** PP
XXXXXXXXXX CS
HSF_DNA-bind 93 llekikrkks 102
ll++i+r+k
WALNUT_00020872-RA 96 LLKNIHRRKP 105
*******985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46785 | 1.45E-36 | 9 | 103 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 7.8E-39 | 10 | 103 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 2.1E-62 | 10 | 103 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 1.3E-31 | 14 | 103 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 6.5E-20 | 14 | 37 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 6.5E-20 | 52 | 64 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 53 | 77 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 6.5E-20 | 65 | 77 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000302 | Biological Process | response to reactive oxygen species | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 390 aa Download sequence Send to blast |
MDDSQGGSNS LPPFLSKTYE MVDDPSTDSV VSWSQCNKSF VVWNPPEFAR VLLPKFFKHN 60 NFSSFIRQLN TYGFRKIDPE QWEFANEDFM RGQPHLLKNI HRRKPVHSHS MQSLQGQGSA 120 SPLSESERQS LKEEIERLKN DRERLLLESQ RNEEERRVYE LQIQRFKERL QQIELRQQNM 180 MSSVAGVLRK PGLALNLVTQ LENHDRKRRL PRISYFRDEA DIEDDQMEAS QSLPRESGAS 240 ASTLTLKMDP FEHLESSLSF WESVVHDVGQ TCVEQNSSVV LDESTSCADS PAISGLQLNV 300 DIRPKSPGID MNSEPATVAA PDSVEPVTSE EQAVVNAATV PTGVNDVFWE QFLTENPGSS 360 EAQEFQSERK ESMAERMKPN LAIMANFSGR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 1e-24 | 7 | 103 | 22 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 1e-24 | 7 | 103 | 22 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 1e-24 | 7 | 103 | 22 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 204 | 209 | DRKRRL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
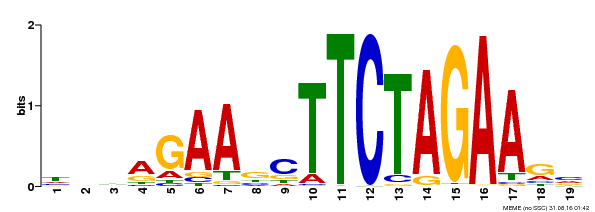
| UniProt | Transcriptional activator that specifically binds DNA sequence 5'-AGAAnnTTCT-3' known as heat shock promoter elements (HSE). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00443 | DAP | Transfer from AT4G18880 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018805848.1 | 0.0 | PREDICTED: heat stress transcription factor A-4c-like | ||||
| Swissprot | O49403 | 1e-117 | HFA4A_ARATH; Heat stress transcription factor A-4a | ||||
| TrEMBL | A0A2I4DFD2 | 0.0 | A0A2I4DFD2_JUGRE; heat stress transcription factor A-4c-like | ||||
| STRING | POPTR_0011s06820.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3568 | 33 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18880.1 | 1e-119 | heat shock transcription factor A4A | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



