 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00024430-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 323aa MW: 35996.8 Da PI: 5.3366 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.6 | 1.8e-19 | 26 | 75 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y+G+r+++ +g+W+AeIrdp++ r++lg+f taeeAa+a++a ++k++g
WALNUT_00024430-RA 26 QYRGIRQRP-WGKWAAEIRDPRK---GVRVWLGTFNTAEEAARAYDAEARKIRG 75
69*******.**********954...4*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.37E-22 | 26 | 85 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 1.48E-33 | 26 | 84 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 1.1E-32 | 26 | 85 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.131 | 26 | 83 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.9E-36 | 26 | 89 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 8.6E-12 | 27 | 38 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.7E-12 | 27 | 75 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 8.6E-12 | 49 | 65 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0070483 | Biological Process | detection of hypoxia | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 323 aa Download sequence Send to blast |
MRSKAIKSVE LNGQAEKSAK RKRKNQYRGI RQRPWGKWAA EIRDPRKGVR VWLGTFNTAE 60 EAARAYDAEA RKIRGKKAKV NFPDEATRAS SKHSVKENLR KPLPKANLHS VQPNLNQNFD 120 SMNYADQDYT MGLMEEKPFT NQYGYLDSIP LNGDVEPKSF VSSGGTTPTS TYFNSDQGSN 180 TFDYSDFAWG EQGPKTPEIS SVLSVSAEGG ESQFLEDASQ FLEDASPRKK LKFDSENVAS 240 VESNTAKTVS VELSAFESQI NYQVPYLEGI WDDSIETLFS GDTTQDVGNP MDLWGFEDDS 300 VMVKLQRKSA SLPLPPGSKT EGN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 2e-21 | 25 | 91 | 13 | 80 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00204 | DAP | Transfer from AT1G53910 | Download |
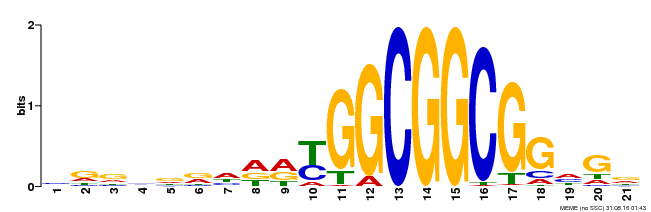
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018809884.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-12-like, partial | ||||
| TrEMBL | A0A2I4DRV7 | 0.0 | A0A2I4DRV7_JUGRE; ethylene-responsive transcription factor RAP2-12-like | ||||
| STRING | VIT_09s0002g00470.t01 | 1e-130 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4808 | 32 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G53910.2 | 1e-56 | related to AP2 12 | ||||



