 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00025554-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1024aa MW: 112718 Da PI: 6.2469 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 66.3 | 5.6e-21 | 61 | 107 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEde++v++v+++G++ W+tIa++++ gR +kqc++rw+++l
WALNUT_00025554-RA 61 KGPWSKEEDEIIVKLVNKYGPKKWSTIAQHLP-GRIGKQCRERWHNHL 107
79******************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 47 | 6.1e-15 | 113 | 155 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE++ l++a++ +G++ W+ + ++ gR+++ +k++w+
WALNUT_00025554-RA 113 KEAWTQEEELALIRAHQIYGNK-WAELTKFLP-GRSDNAIKNHWN 155
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 3.984 | 1 | 55 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 8.7E-11 | 36 | 67 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 33.31 | 56 | 111 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.83E-31 | 58 | 154 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.2E-19 | 60 | 109 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.2E-19 | 61 | 107 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.08E-17 | 63 | 107 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-27 | 68 | 115 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.978 | 112 | 162 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.1E-14 | 112 | 160 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.4E-13 | 113 | 156 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.14E-11 | 115 | 155 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-20 | 116 | 162 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1024 aa Download sequence Send to blast |
MQVYLFVKKL WKVTRQSILL KMGLVMVVRK CDLYTAECFK ERTDVQCLHR WQKVLNPELV 60 KGPWSKEEDE IIVKLVNKYG PKKWSTIAQH LPGRIGKQCR ERWHNHLNPA INKEAWTQEE 120 ELALIRAHQI YGNKWAELTK FLPGRSDNAI KNHWNSSVKK KLDSYLASGL LAQFPALPHI 180 GHQNQHMHSS SSRMQGSGDD SGPKGIETQE TSECSQESTV AGCYKSSSDM TNAVLHASEE 240 FPLTRESGSA KEQSSSPPSC SEQYYTSLED VDFSMPNITC EADCSDKFLE QNFSHDAGTS 300 AGGDHQFNIH AIPNISSLEV EQESSQLKTH CIGTNESHET MDVPCQTSAE LRASTSTGCI 360 TMGSYKLENM RIPDDECCRI LFSEALNDGF FSGSHTKVSN IVPLGECTDS FCCQSSNCII 420 PEADGTSAVQ SYCPPRSDLV GSPCSQSFRF LSSLISVDCA TLLYGDEPNQ LFETQEEGFV 480 TGAHEGFIYT NDFTNSPSNN GTDNAGMQEQ PDVKETSKLV PVNTFGSGSD ITDTCPSVGE 540 RLHVHLEQQD AGALCYEPPR FPSLDIPFLS CDLIQSGSDT QQEYSPLGIR QLMMSSMNCI 600 TPFRLWDSPS RDDSPDAFLK SAAKTFTGTP SILKKRHRDL FSPLSDRRSD KKLEIDMTSS 660 LSRHFAHLDV VFDEISTQNA PFSPSSNRKR NFGASPMDKE NTDCAFKGAE GKGRDGTSSF 720 DDRNLEKNSD DSNLQGNTKQ PTADVDAKTK VQQSSGVLVE HNMNDLLLHS PDQVCSKADR 780 LLRSSSRTPR NKQSRSLAAA SDRGISNHLS SGNPCAHFRS PTLCGKKNES HSVAVTCTQS 840 ASSSAPLETA GHNVRNDAGV ETFGIFGGTP FKRSIESPSA WKSPWFINSF LPGPRVDTEI 900 TIEDIGYFMS PGDRSYDAIG LMKHISEHSA AAYANAQEVL GNETPKTLKK GKCNNHGSRD 960 QENSRVPYSH HGNRSHMAPT VLTECRTLDF SECGTPGKGT ENGKPSNAVS LSSSPSSYLL 1020 KNCR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-61 | 36 | 162 | 33 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-61 | 36 | 162 | 33 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00466 | DAP | Transfer from AT4G32730 | Download |
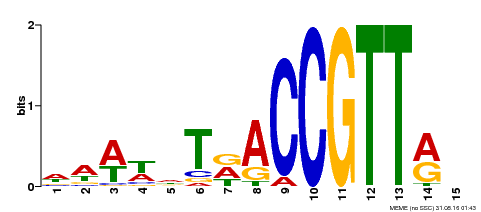
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018811761.1 | 0.0 | PREDICTED: myb-related protein 3R-1-like isoform X2 | ||||
| TrEMBL | A0A2I4DX84 | 0.0 | A0A2I4DX84_JUGRE; myb-related protein 3R-1-like isoform X2 | ||||
| STRING | cassava4.1_024255m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3727 | 33 | 62 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32730.2 | 0.0 | Homeodomain-like protein | ||||



