 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004493567.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 350aa MW: 38574.8 Da PI: 8.4943 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 133.6 | 6.4e-42 | 57 | 133 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+CqvegC++dl++ak y++rhkvC +hsk+p+v+v+ l+qrfCqqCsrfh+l efD++krsCrrrLa+hnerrrk+
XP_004493567.1 57 RCQVEGCQVDLTDAKPYYSRHKVCAMHSKSPTVTVAALQQRFCQQCSRFHQLVEFDQGKRSCRRRLAGHNERRRKPP 133
6**************************************************************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.2E-33 | 51 | 119 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.573 | 55 | 132 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.96E-39 | 56 | 137 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.0E-31 | 58 | 131 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:2000025 | Biological Process | regulation of leaf formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 350 aa Download sequence Send to blast |
MGSSNSSSED SSLNGLKFGQ TIYFEDVAAI PPPPPSIATS SKKGGRCASL QQSQPPRCQV 60 EGCQVDLTDA KPYYSRHKVC AMHSKSPTVT VAALQQRFCQ QCSRFHQLVE FDQGKRSCRR 120 RLAGHNERRR KPPPSSLLTS RFARLSSSVF GNSGRGSSFL MEFNSHAKLS MRNPLPSPRS 180 SELFPGNQTT TLGWHWPGNS ESPSDNLFLQ GSVGGTSFPG TRHPPEESYT GVTDSSCARS 240 LLSNQTWGSR NTEPSLRLNN MLNFNETPMT QLATSSHSVA IHQLPNNSFC YKDIDPGNIS 300 HEVVPDLGLG HMSQPLDSQL SGELDLSQQG RRHYMDPEHS RAYESSQWSL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-30 | 58 | 131 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00307 | DAP | Transfer from AT2G42200 | Download |
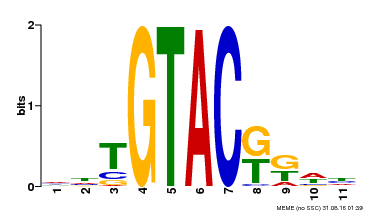
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004493567.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC157891 | 4e-89 | AC157891.3 Medicago truncatula chromosome 7 clone mte1-1i2, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004493567.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A1S2XRU3 | 0.0 | A0A1S2XRU3_CICAR; squamosa promoter-binding-like protein 9 | ||||
| STRING | XP_004493567.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4208 | 33 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42200.1 | 2e-67 | squamosa promoter binding protein-like 9 | ||||



