 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004496891.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 809aa MW: 93391.2 Da PI: 7.0699 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 88.9 | 7.4e-28 | 48 | 135 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHelap 91
+fY+eYAk++GF++++++s++sk+++e+++++f Cs++g + e+++ ++r+ ++++t+Cka+++vkk+ dgkw ++++ ++HnHel p
XP_004496891.1 48 SFYQEYAKSMGFTTSIKNSRRSKKTKEFIDAKFACSRYGVTPESDSG---SSRRPSVKKTDCKACMHVKKRPDGKWTIHEFIKDHNHELLP 135
5****************************************999888...7788889*******************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 5.7E-25 | 48 | 135 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 8.4E-30 | 255 | 348 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 9.285 | 535 | 571 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 6.2E-7 | 537 | 570 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 3.4E-8 | 546 | 573 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 809 aa Download sequence Send to blast |
MSDAVNEEDR RGGAPVASPK RDVAFFEGDR DFEPPNGIEF ESHEAAYSFY QEYAKSMGFT 60 TSIKNSRRSK KTKEFIDAKF ACSRYGVTPE SDSGSSRRPS VKKTDCKACM HVKKRPDGKW 120 TIHEFIKDHN HELLPALAYH FRIHRNVKLA EKNNIDILHA VSERTKKMYV EMSRQSGGCQ 180 NPESLVGDTN YQFDRGQYLS LDEGDAQVML EYFKHIQKEN PNFFYSIDLN EEQRLRNLFW 240 VDAKSINDYL SFNDVVSFDT TYVKSNDKLP FAPFIGVNHH SQPILLGCAL VADETKPTFV 300 WLLKTWLRAM GGQAPKVIVT DQDKALKAAI EEVFPNVRHC FSLWHILEKI PENLSFVIKQ 360 YKNFLPKFNN CIFKSWTDEQ FDMRWWEMVT IFELHDDVWF HSLYEDRKKW VPTYMGDVFL 420 AGMSTSQRSE SMNSFFDKYI HKKITLKEFV KQYGLILQNR YDEEAIADFD TLHKQPALKS 480 PSPWEKQMST IYTHTIFKKF QIEVLGVAGC QSRIEVGDGN VAKFIVQDYE KDEEFLVTWN 540 ELSSEVSCFC RLFEYKGFLC RHALSVLQRC GCSSVPSHYI MKRWTKDAKI REHIADRTRR 600 IQTRVQRYND LCKRAIELSE EGSLSEESYN VAIRTLIDSL KNCVLVNNSN GNGAETGNNG 660 YSLREAEQNQ VTLASKPSKK RNTTRKRKVQ QEQNPILVDA QDSLQQMDNL SSDAMTLNGY 720 YGTQQNVQGL QVQLNLMEPP HDGYYVNQQS MQGLGPLNSM APSHDGYFGT QQSIHGMGGQ 780 LEYRPTTPFG YSLQDEPDQH FHSNNSRNT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
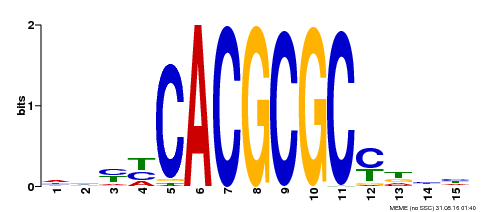
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004496891.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC235177 | 0.0 | AC235177.1 Glycine max strain Williams 82 clone GM_WBb0007H11, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004496891.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A1S2Y0L0 | 0.0 | A0A1S2Y0L0_CICAR; protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| STRING | XP_004496891.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF40 | 30 | 530 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||



