 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004500737.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 334aa MW: 38553.2 Da PI: 9.6537 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.4 | 6.8e-13 | 282 | 328 | 2 | 47 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.TTS-HHHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmg.kgRtlkqcksrwqky 47
+rW++ E+e l av++lG g+W++I +t +Rt+ ++k++w++
XP_004500737.1 282 KRWSELEEETLRTAVRKLGEGNWTAILNTYTfDNRTGVDLKDKWRNM 328
69*******************************************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.669 | 273 | 333 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.58E-13 | 278 | 329 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-9 | 280 | 331 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-13 | 282 | 328 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.5E-10 | 282 | 328 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 1.34E-17 | 282 | 330 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 334 aa Download sequence Send to blast |
MEDDVRWCLE FLGGKSSVDI SEVNKRFLRM SVSDNCSRLK KTILLQTLQD ELYSKASTSE 60 WMLEALEVME TLLLSEDSCS NQITATMKDA YCAIAVECTI KYLEVETCNP AYRQAVERIW 120 KGRVQKMEPS SEREGSFLFS PELKRWKTII VVSLLNPTYV DMFASITHPR RDALKKLKLY 180 LDEALQNLAL SRNHQTEIQK DNETSLKGKV TTTGIDDANL TKTVTVYDVC DSSFREPNRA 240 ARTHKGGTSN KTRITLISPK KVKISPLKKY EPKKIFRRRK PKRWSELEEE TLRTAVRKLG 300 EGNWTAILNT YTFDNRTGVD LKDKWRNMER SGRQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
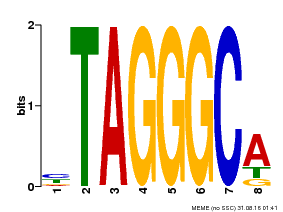
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004500737.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004500737.1 | 0.0 | uncharacterized protein LOC101498622 | ||||
| TrEMBL | A0A1S2Y7S5 | 0.0 | A0A1S2Y7S5_CICAR; uncharacterized protein LOC101498622 | ||||
| STRING | XP_004500737.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2824 | 32 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 2e-15 | TRF-like 5 | ||||



