 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004502338.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 590aa MW: 66937.8 Da PI: 6.3501 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 74.6 | 1.4e-23 | 199 | 252 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++W+ +LH++Fv+av+ + G +k Pk+il+lm+v+ Lt+e+v+SHLQkYRl
XP_004502338.1 199 KARVVWSVDLHQKFVKAVNLI-GFDKVGPKKILDLMNVPWLTRENVASHLQKYRL 252
68*****************99.********************************8 PP
| |||||||
| 2 | Response_reg | 83.9 | 4.9e-28 | 19 | 127 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaG 93
vl+vdD+p+ +++l+++l+k +y ev+++ +++al+ll+e++ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge + + + ++ G
XP_004502338.1 19 VLVVDDDPTWLKILEKMLKKCNY-EVTTCCLARQALNLLRERKdgYDIVISDVNMPDMDGFKLLEHVGLEM-DLPVIMMSVDGETSRVMKGVQHG 111
89*********************.***************999999**********************6644.8********************** PP
ESEEEESS--HHHHHH CS
Response_reg 94 akdflsKpfdpeelvk 109
a d+l Kp+ ++el++
XP_004502338.1 112 ACDYLLKPIRMKELRN 127
*************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 9.4E-146 | 2 | 559 | IPR017053 | Response regulator B-type, plant |
| SuperFamily | SSF52172 | 6.83E-37 | 15 | 145 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 1.1E-44 | 16 | 153 | No hit | No description |
| SMART | SM00448 | 9.6E-33 | 17 | 129 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 43.657 | 18 | 133 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 4.9E-25 | 19 | 128 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 8.78E-31 | 20 | 133 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.7E-27 | 196 | 257 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 7.17E-17 | 197 | 256 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.5E-21 | 199 | 252 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.6E-7 | 201 | 251 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 590 aa Download sequence Send to blast |
MDNGCFTSPR RDFPNGLRVL VVDDDPTWLK ILEKMLKKCN YEVTTCCLAR QALNLLRERK 60 DGYDIVISDV NMPDMDGFKL LEHVGLEMDL PVIMMSVDGE TSRVMKGVQH GACDYLLKPI 120 RMKELRNIWQ HVLRKRIHEV REFENFESFE SIHLMRNGSE MSEEANMFAL EDMTSTKKRK 180 DVDNKHDDKE FSDPSSSKKA RVVWSVDLHQ KFVKAVNLIG FDKVGPKKIL DLMNVPWLTR 240 ENVASHLQKY RLYLSRLQKD NEQKSSSSGM KHSDLPSKDV GSFGFPNSAN MKHSDLPSKD 300 VGSYGFPNSA NKQQNDVPID RYNYLEGTIQ HKNMETKSHE GIPKVTVPHS TTAEKVRAIT 360 GNITDAGITH NRPFAPLDSE GKCAVIDCTL STQYSWTEIP KRQLKEEQES LVHLEDNCNM 420 LPLQGKKHHI QVDQSQSITS ISSTPSTKEQ EVDAIIKTKH LLADYKKDYT SSVSSKRSVV 480 DTFPFQPGSL MMNDQPSQPI STSNFGLKTQ SSNLGCIPDL ESYQRSLLLV GEAASAPLDE 540 DLYFYWHNMN FGQSNIGMSE YYDPGLLTEV PTHLYDSTDY SVIDQGLFIA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-16 | 195 | 257 | 1 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
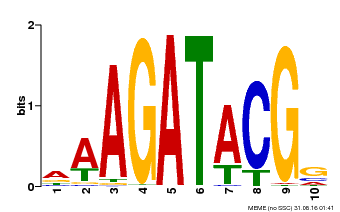
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004502338.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT140505 | 0.0 | BT140505.1 Medicago truncatula clone JCVI-FLMt-22H12 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004502338.1 | 0.0 | two-component response regulator ORR26 | ||||
| TrEMBL | A0A1S2YBE1 | 0.0 | A0A1S2YBE1_CICAR; Two-component response regulator | ||||
| STRING | XP_004502338.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3999 | 33 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-121 | response regulator 11 | ||||



