 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004506004.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 486aa MW: 53565.2 Da PI: 7.9558 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.5 | 6.1e-17 | 158 | 207 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
XP_004506004.1 158 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 207
78*******************......55************************997 PP
| |||||||
| 2 | AP2 | 42.3 | 1.9e-13 | 250 | 298 | 1 | 53 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkl 53
s+y+GV+ +k grW+A+ + +k+++lg f+t+ eAa+a+++a+ k
XP_004506004.1 250 SKYRGVTLHK-CGRWEARMGQF--L-GKKYIYLGLFDTEVEAARAYDKAAIKN 298
89********.7******5553..2.26**********99**********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.57E-15 | 158 | 216 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 5.6E-9 | 158 | 207 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.6E-31 | 159 | 221 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.2E-16 | 159 | 215 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.23E-10 | 159 | 214 | No hit | No description |
| PROSITE profile | PS51032 | 17.411 | 159 | 215 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 6.54E-16 | 250 | 309 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 3.01E-24 | 250 | 310 | No hit | No description |
| Pfam | PF00847 | 1.2E-8 | 250 | 298 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.6E-28 | 251 | 314 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 7.2E-15 | 251 | 308 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 15.33 | 251 | 308 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 486 aa Download sequence Send to blast |
MLDLNLNDAN STDSTQNNDS PMISEKFPQT SWNQMVESGT SNSSIVNAGG SSNNANDEDS 60 CSTRADDAFT FNFDILKVEG SNSNDIVTKE LFPDSGGGDE RWSKNLQWQN TSSFPTRNGT 120 SVDLSFKQLC GNEEMKMVQV QPQQQAKKSR RGPRSRSSQY RGVTFYRRTG RWESHIWDCG 180 KQVYLGGFDT AHAAARAYDR AAIKFRGVGA DINFNLNDYD DDLQQAKNLS KEEFVHILRR 240 QSTGFSRGSS KYRGVTLHKC GRWEARMGQF LGKKYIYLGL FDTEVEAARA YDKAAIKNNA 300 REAVTNFEPS TYEGELKSTA IIEGGSQNLD LNLGIATPGT GPKENLGKFH FPSIPYTMQA 360 GRSSMMETNV SGIGNPSLKR LLWNGMSPAF FPNEERAESI SKDPSKGLPN WGWQTHGQVT 420 ATPLPAVSIA ASSGFSISPP SFPSTAIFPP RSMNSIPQSL YFTSSNAPSS NALQNYYQVN 480 PPQAPP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Regulates negatively the transition to flowering time and confers flowering time delay. {ECO:0000250, ECO:0000269|PubMed:14555699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
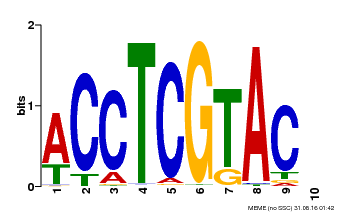
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004506004.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172a-2/EAT. {ECO:0000269|PubMed:14555699}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027191944.1 | 0.0 | ethylene-responsive transcription factor RAP2-7 | ||||
| Swissprot | Q9SK03 | 1e-118 | RAP27_ARATH; Ethylene-responsive transcription factor RAP2-7 | ||||
| TrEMBL | A0A3Q7XEL8 | 0.0 | A0A3Q7XEL8_CICAR; ethylene-responsive transcription factor RAP2-7 | ||||
| STRING | XP_004506004.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2807 | 34 | 76 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 1e-134 | related to AP2.7 | ||||



