 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004509044.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 288aa MW: 32128.2 Da PI: 8.2062 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.9 | 1.8e-18 | 142 | 196 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ ++tk+q Lee F+++++++ ++++ LA++l+L rqV vWFqNrRa+ k
XP_004509044.1 142 RKKLRLTKDQSAMLEENFKQHSTLNPKQKQALARELNLLPRQVEVWFQNRRARTK 196
788899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 124.2 | 6e-40 | 142 | 231 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
+kk+rl+k+q+++LEe+F+++++L+p++K++lareL+l prqv+vWFqnrRARtk+kq+E+d+e+Lk+++++l++en+rL+kev+eL+ +l
XP_004509044.1 142 RKKLRLTKDQSAMLEENFKQHSTLNPKQKQALARELNLLPRQVEVWFQNRRARTKLKQTEVDCEFLKKCCETLTDENRRLQKEVQELK-AL 231
69*************************************************************************************9.55 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 16.908 | 138 | 198 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.7E-15 | 140 | 202 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 5.13E-16 | 142 | 199 | No hit | No description |
| Pfam | PF00046 | 8.1E-16 | 142 | 196 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-17 | 142 | 196 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 6.42E-17 | 142 | 206 | IPR009057 | Homeodomain-like |
| PROSITE pattern | PS00027 | 0 | 173 | 196 | IPR017970 | Homeobox, conserved site |
| Pfam | PF02183 | 1.2E-10 | 198 | 232 | IPR003106 | Leucine zipper, homeobox-associated |
| SMART | SM00340 | 3.7E-26 | 198 | 241 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 288 aa Download sequence Send to blast |
MGLDQDASSN NSGLPLILGL ALTLSQQETI TPPPSNKITK PFNNSSNFIT CSTSNNHNNN 60 LQAELPSLTL GLSYPKNVNN NKVYYEDPLD FSTQTSPHHS VVSSFSSGRV KRERDVSSEE 120 VEAIERVCSR VSDEEEDAVA ARKKLRLTKD QSAMLEENFK QHSTLNPKQK QALARELNLL 180 PRQVEVWFQN RRARTKLKQT EVDCEFLKKC CETLTDENRR LQKEVQELKA LKLTQPLYMP 240 MPAATLTMCP SCERLGGVSG GGASNKTTFS MAPKPHFYNP FTNSSAAC |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 190 | 198 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00477 | DAP | Transfer from AT4G37790 | Download |
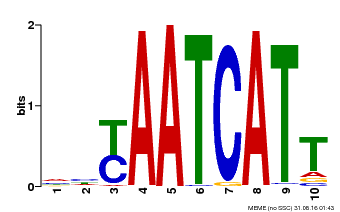
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004509044.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004509044.1 | 0.0 | homeobox-leucine zipper protein HAT22 | ||||
| Swissprot | P46604 | 9e-91 | HAT22_ARATH; Homeobox-leucine zipper protein HAT22 | ||||
| TrEMBL | A0A1S2YS47 | 0.0 | A0A1S2YS47_CICAR; homeobox-leucine zipper protein HAT22 | ||||
| STRING | XP_004509044.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1144 | 33 | 107 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37790.1 | 1e-90 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



