 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004510747.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 331aa MW: 36277 Da PI: 9.2683 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43 | 1.1e-13 | 58 | 102 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT +E+e++++a +++ + Wk+I + +g +t q++s+ qky
XP_004510747.1 58 RESWTDQEHEKFLEALQLFDRD-WKKIEAFVG-SKTVIQIRSHAQKY 102
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 8.97E-17 | 52 | 108 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.486 | 53 | 107 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 4.2E-18 | 56 | 105 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 8.1E-7 | 56 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.9E-11 | 57 | 105 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.2E-11 | 58 | 102 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.99E-8 | 60 | 103 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MVSVNPNNPA QAFYFFDPSN MALPGANHLP PPTSAAVSTT VDDPNKKIRK PYTITKSRES 60 WTDQEHEKFL EALQLFDRDW KKIEAFVGSK TVIQIRSHAQ KYFLKVQKNG TSEHVPPPRP 120 KRKAAHPYPQ KAPKNAPTAS QVMGPLQSSP AFIEPAYIYN TDSSSVLGTP VTSVPLSSWN 180 CNAKPPASVP QVTKDDTGWT GSGQAVPLNC CHSSSNESTH PALPSSKGVN QGNLGKPLNV 240 MPDFSQVYNF IGSVFDPNST NHLQRLKQMD PINVETALLL MRNLSINLTS PEFEDHKKLL 300 SSYDADCEKA KLANPSSKPM TVKSENPVLS A |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00423 | DAP | Transfer from AT4G01280 | Download |
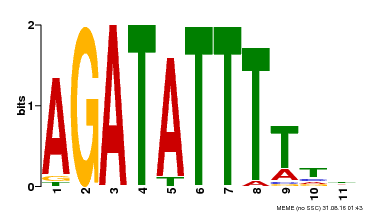
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004510747.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF468481 | 0.0 | EF468481.1 Medicago truncatula MYB transcription factor MYB40 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004510747.1 | 0.0 | protein REVEILLE 3 | ||||
| Swissprot | C0SVG5 | 1e-108 | RVE5_ARATH; Protein REVEILLE 5 | ||||
| TrEMBL | A0A1S2YVV0 | 0.0 | A0A1S2YVV0_CICAR; protein REVEILLE 3 | ||||
| STRING | XP_004510747.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1119 | 33 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01280.1 | 1e-96 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



