 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004510857.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 807aa MW: 88960.1 Da PI: 6.1868 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.2 | 4.5e-21 | 117 | 172 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
XP_004510857.1 117 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 172
688999***********************************************999 PP
| |||||||
| 2 | START | 210.2 | 7.6e-66 | 319 | 543 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetl 82
ela++a++elvk+a+ +ep+W +s e++n +e++++f++ + + +ea+r++g+v+ ++ lve+l+d++ +W e+++ + +t+
XP_004510857.1 319 ELALAAMDELVKMAQTSEPLWIRSIeggrEILNHEEYMRTFTPCIGlrpngFVSEASRETGMVIINSLALVETLMDSN-RWIEMFPciiaRTSTT 412
5899**************************************9999********************************.**************** PP
EEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-E CS
START 83 evissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehv 170
evis+g galqlm+ael +lsplvp R++ f+R+++q+ +g+w++vdvS+ds ++++ +s+v +++lpSg+++++++ng+skvtwveh+
XP_004510857.1 413 EVISNGingtrnGALQLMQAELHVLSPLVPvREVNFLRFCKQHAEGVWAVVDVSIDSIRENSGAPSFVNCRKLPSGCVVQDMPNGYSKVTWVEHA 507
**************************************************************9******************************** PP
E--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 171 dlkgrlphwllrslvksglaegaktwvatlqrqcek 206
+++++++h+l+r+l++sg+ +ga +wv tlqrqce+
XP_004510857.1 508 EYEENQVHQLYRPLLSSGMGFGATRWVVTLQRQCEC 543
**********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.54E-20 | 102 | 174 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 9.8E-22 | 106 | 174 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 114 | 174 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.9E-18 | 115 | 178 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.63E-18 | 116 | 174 | No hit | No description |
| Pfam | PF00046 | 1.4E-18 | 117 | 172 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 149 | 172 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.721 | 310 | 546 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.65E-34 | 312 | 543 | No hit | No description |
| CDD | cd08875 | 6.08E-127 | 314 | 542 | No hit | No description |
| Pfam | PF01852 | 2.1E-57 | 319 | 543 | IPR002913 | START domain |
| SMART | SM00234 | 1.8E-48 | 319 | 543 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.2E-23 | 571 | 800 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 807 aa Download sequence Send to blast |
MSFGGFVENN SGGGSVRNIA AEISYNNNQR MSFGSISHPR LVTTPTLAKS MFNSPGLSLA 60 LQTNIDGQED VNRSMHENFE QNGLRRSREE EQSRSGSDNL DGVSGDEQDA DDKPPRKKRY 120 HRHTPQQIQE LEALFKECPH PDEKQRLELS KRLCLETRQV KFWFQNRRTQ MKTQLERHEN 180 SLLRQENDKL RAENMSIRDA MRNPICSNCG GPAMIGEISL EEQHLRIENA RLKDELDRVC 240 ALAGKFLGRP ISTLPNSSLE LGVGGNNGFN GMNNVSSTLP DFGVGMSNNP LAIVSPSTRQ 300 TTPLVTGFDR SVERSMFLEL ALAAMDELVK MAQTSEPLWI RSIEGGREIL NHEEYMRTFT 360 PCIGLRPNGF VSEASRETGM VIINSLALVE TLMDSNRWIE MFPCIIARTS TTEVISNGIN 420 GTRNGALQLM QAELHVLSPL VPVREVNFLR FCKQHAEGVW AVVDVSIDSI RENSGAPSFV 480 NCRKLPSGCV VQDMPNGYSK VTWVEHAEYE ENQVHQLYRP LLSSGMGFGA TRWVVTLQRQ 540 CECLAILMSS AAPSRDHSAI TAGGRRSMLK LAQRMTNNFC AGVCASTVHK WNKLSPGNVD 600 EDVRVMTRKX XXDPGEPPGI VLSAATSVWL PVSPQRLFDF LRDERLRSEW DILSNGGPMQ 660 EMAHIAKGQD HGNCVSLLRA SAMNSNQSSM LILQETCIDE AGSLVVYAPV DIPAMHVVMN 720 GGDSAYVALL PSGFAVVPDG PGSRGPENET TTNGGETRVS GSLLTVAFQI LVNSLPTAKL 780 TVESVETVNN LISCTVQKIK AALQCES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
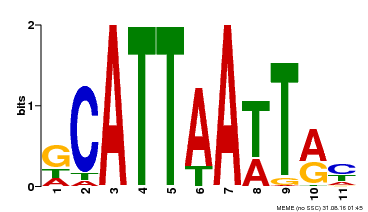
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004510857.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004510857.1 | 0.0 | homeobox-leucine zipper protein ANTHOCYANINLESS 2-like isoform X2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A1S2YW50 | 0.0 | A0A1S2YW50_CICAR; homeobox-leucine zipper protein ANTHOCYANINLESS 2-like isoform X2 | ||||
| STRING | XP_004510857.1 | 0.0 | (Cicer arietinum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



