 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004511225.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 282aa MW: 32188.8 Da PI: 6.1786 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.3 | 1.3e-18 | 76 | 129 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rqV +WFqNrRa++k
XP_004511225.1 76 KKRRLNMEQVKTLEKSFELGNKLEPERKMQLARALGLQPRQVAIWFQNRRARWK 129
456899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 133.6 | 6.9e-43 | 75 | 165 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ eqvk+LE+sFe +kLeperK++lar+Lglqprqva+WFqnrRAR+ktkqlEkdy++Lkr+yd++k++n++L++++++L++e+
XP_004511225.1 75 EKKRRLNMEQVKTLEKSFELGNKLEPERKMQLARALGLQPRQVAIWFQNRRARWKTKQLEKDYDVLKRQYDSIKADNDALQAQNQKLQTEI 165
69**************************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.2E-19 | 70 | 133 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 71 | 131 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-18 | 74 | 135 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 6.2E-16 | 76 | 129 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.13E-15 | 76 | 132 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-19 | 78 | 138 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 8.7E-6 | 102 | 111 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 106 | 129 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 8.7E-6 | 111 | 127 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 6.0E-17 | 131 | 171 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 282 aa Download sequence Send to blast |
MAFFPTNFML QTSHQDDHQT PPSLNSIITS CAPQEYHGGG VSFLGKRSMS FSGIELGEEA 60 NVEEELSDDG SQLGEKKRRL NMEQVKTLEK SFELGNKLEP ERKMQLARAL GLQPRQVAIW 120 FQNRRARWKT KQLEKDYDVL KRQYDSIKAD NDALQAQNQK LQTEILALKS REPTESINLN 180 KETEGSSSNR SENSSEIKLD ISTRTQAIDS PLSTQQTSRN LFPSSSRPTG VAQLFQTNSR 240 QDHIQCQKIN DHMVKEESLS NMFCGIDDQS GLWPWLEHQH FN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 123 | 131 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may act in the sucrose-signaling pathway. {ECO:0000269|PubMed:11292072}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
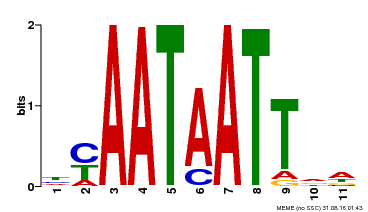
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004511225.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT051961 | 0.0 | BT051961.1 Medicago truncatula clone MTYF9_FA_FB_FC1G-H-4 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004511225.1 | 0.0 | homeobox-leucine zipper protein ATHB-13-like | ||||
| Swissprot | Q8LC03 | 1e-115 | ATB13_ARATH; Homeobox-leucine zipper protein ATHB-13 | ||||
| TrEMBL | A0A1S2YWW9 | 0.0 | A0A1S2YWW9_CICAR; homeobox-leucine zipper protein ATHB-13-like | ||||
| STRING | XP_004511225.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1271 | 33 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 1e-105 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



