 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004512346.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 650aa MW: 73714.1 Da PI: 5.9497 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 152.4 | 3.4e-47 | 45 | 194 | 1 | 145 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
+g+g+k+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGw+v++DGttyr ++ p a+ +gs+a +s+es+l+
XP_004512346.1 45 TGKGKKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWIVDADGTTYR-QCPP---ASNMGSFAARSVESQLS 135
589*******************************************************************.3444...5899************* PP
DUF822 96 .sslkssalaspvesysa.spksssfpspssldsislasa.......asllpvlsvlsl 145
sl++++++ +e++ + +++ ++ +sp+s+ds+ +a+ as++p+ sv++l
XP_004512346.1 136 gGSLRTCSVQETLENQPPvVLRIDECLSPASIDSVVIAERdskngkyASVSPINSVDCL 194
99999999888887776516799**************9888888888778888877775 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 3.4E-48 | 46 | 194 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 3.16E-156 | 212 | 646 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.5E-168 | 215 | 645 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 1.0E-84 | 237 | 617 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 252 | 266 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 273 | 291 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 295 | 316 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 388 | 410 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 461 | 480 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 495 | 511 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 512 | 523 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 530 | 553 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.8E-56 | 568 | 590 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 650 aa Download sequence Send to blast |
MKSINEDASV LNLDPQSDHS SDYLAQPQPR RPRGFAAAAM GTNSTGKGKK EREKEKERTK 60 LRERHRRAIT SRMLAGLRQY GNFPLPARAD MNDVLAALAR EAGWIVDADG TTYRQCPPAS 120 NMGSFAARSV ESQLSGGSLR TCSVQETLEN QPPVVLRIDE CLSPASIDSV VIAERDSKNG 180 KYASVSPINS VDCLEADQLM QDIHSGVHES DFNCTPYVPV YIKLPAGIIN KFCQLIDPEG 240 IRQELIHIKS LNIDGVVVDC WWGIVEGWSP QKYVWSGYRE LFNIIREFKL NLQVVMAFHE 300 CGGNDSSDAL ISLPQWVLDI GKENQDIFFT DREGRRNTEC LSWGIDKERV LKGRTGIEVY 360 FDMMRSFRTE FDDLFAEGLI YAVEIGLGAS GELKYPSFSE RMGWRYPGIG EFQCYDKYLQ 420 HSLRRAAKLR GHSFWARGPD NAGHYNSMPH ETGFFCERGD YDNYYGRFFL HWYSQTLTDH 480 ADNVLSLASL AFEETKIIVK VPAVYWWYKS PSHAAELTAG YHNPTNQDGY SPVFEVLRKH 540 AVTMKFVCLG FNLSSQEANE SLVDPEGLSW QALNSAWERG LITAGENALF GYDRERYKRL 600 VEMAKPRNDP DHRHFSFFVY QQPSLLQGNV CLSELDFFIK CMHGEMTGDL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-127 | 217 | 607 | 11 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
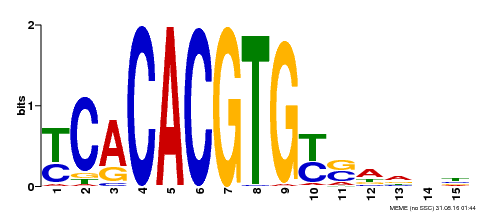
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004512346.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004512346.1 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A1S2YZF6 | 0.0 | A0A1S2YZF6_CICAR; Beta-amylase | ||||
| STRING | XP_004512346.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



