 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_004515249.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Cicereae; Cicer
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 722aa MW: 80172.3 Da PI: 6.4664 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.9 | 4e-19 | 28 | 83 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q+++Le++F+++++p++++r +L+++l L rq+k+WFqNrR+++k
XP_004515249.1 28 KKRYHRHTANQIQRLEAMFKECPHPDEKQRLQLSRELALAPRQIKFWFQNRRTQMK 83
678899***********************************************998 PP
| |||||||
| 2 | START | 183.7 | 1e-57 | 227 | 450 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEE CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla....kaetl 82
+a a++elv++ + ++p+W ks + +n + + ++f+++++ ++ea+r+sgvv+m++ +lve ++d + +W e ++ a t+
XP_004515249.1 227 IATNAMDELVRLLQTNDPLWIKSNtdgkDVLNLEAYERIFPKTNNrlknpnLRIEASRDSGVVIMNSLTLVEMFMDPN-KWMELFPtivtMARTI 320
56789********************999999**********9999*********************************.999999999999**** PP
EEECTT.....EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE CS
START 83 evissg.....galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvd 171
evissg g lqlm+ elq+lsplvp R+f+f+Ry+ q ++g w+ivdvS + + +++ + R+++lpSg+ i++++ng+skvtw+ehv+
XP_004515249.1 321 EVISSGimghsGSLQLMYEELQVLSPLVPiREFYFLRYCHQIEQGLWAIVDVSYEFPHDNQ-FTPQFRCHRLPSGCFIQDMPNGYSKVTWIEHVE 414
************************************************************9.79999**************************** PP
--SSXX.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 172 lkgrlp.hwllrslvksglaegaktwvatlqrqcek 206
+++ p h l+r ++ sg+a+ga +w+ tlqr ce+
XP_004515249.1 415 IEDKNPiHRLYRNVIYSGVAFGAERWLTTLQRMCER 450
**********************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.6E-20 | 4 | 85 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-21 | 13 | 87 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.686 | 25 | 85 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.8E-17 | 26 | 89 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.40E-17 | 27 | 86 | No hit | No description |
| Pfam | PF00046 | 9.8E-17 | 28 | 83 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 60 | 83 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 52.072 | 217 | 453 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.26E-39 | 218 | 451 | No hit | No description |
| CDD | cd08875 | 3.18E-118 | 221 | 449 | No hit | No description |
| SMART | SM00234 | 1.8E-42 | 226 | 450 | IPR002913 | START domain |
| Pfam | PF01852 | 1.0E-49 | 227 | 450 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 1.3E-7 | 263 | 416 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 3.97E-20 | 483 | 647 | No hit | No description |
| SuperFamily | SSF55961 | 3.97E-20 | 674 | 711 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 722 aa Download sequence Send to blast |
MEYHSGGSGS PGDHHHHHDG SSDSQRRKKR YHRHTANQIQ RLEAMFKECP HPDEKQRLQL 60 SRELALAPRQ IKFWFQNRRT QMKAQHERAD NCALRAENDK IRCENIAIRE ALKNVICPSC 120 GGPPINDDCY FDEQKLRIEN AQLKEELDRV SSIAAKYIGR PISQLPPVQP IHISSLDLSM 180 SSYGGQGLGG PSLDLDLLPG SSSSAMQNVP YQPACLSDME KSLMSDIATN AMDELVRLLQ 240 TNDPLWIKSN TDGKDVLNLE AYERIFPKTN NRLKNPNLRI EASRDSGVVI MNSLTLVEMF 300 MDPNKWMELF PTIVTMARTI EVISSGIMGH SGSLQLMYEE LQVLSPLVPI REFYFLRYCH 360 QIEQGLWAIV DVSYEFPHDN QFTPQFRCHR LPSGCFIQDM PNGYSKVTWI EHVEIEDKNP 420 IHRLYRNVIY SGVAFGAERW LTTLQRMCER LACLMVSGNS TRDLGGVIPS PEGXXXXXXX 480 XXXMVTNFCA SISASASHRW TTLSALNEIG VRVTVRKSTD PGQPNGVVLS AATTIWLPLP 540 PQTVFNFFKD ERKRSQWDVL SNGNAVQEVA HIANGSHPGN CISVLRAFNT SQNNMLILQE 600 SCVDSSGSLV VYCPVDLPAI NIAMSGEDPS YIPLLPSGFT ITSDGNQNDN NNNNNNNNVG 660 DGASTSSNTN LGGGSLITVA FQIMVSSLPS AKLNMESVAT VNGLIGETVQ HIKAALNCPS 720 SG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which acts as positive regulator of drought stress tolerance. Can transactivate CIPK3, NCED3 and ERECTA (PubMed:18451323). Transactivates several cell-wall-loosening protein genes by directly binding to HD motifs in their promoters. These target genes play important roles in coordinating cell-wall extensibility with root development and growth (PubMed:24821957). Transactivates CYP74A/AOS, AOC3, OPR3 and 4CLL5/OPCL1 genes by directly binding to HD motifs in their promoters. These target genes are involved in jasmonate (JA) biosynthesis, and JA signaling affects root architecture by activating auxin signaling, which promotes lateral root formation (PubMed:25752924). Acts as negative regulator of trichome branching (PubMed:16778018, PubMed:24824485). Required for the establishment of giant cell identity on the abaxial side of sepals (PubMed:23095885). May regulate cell differentiation and proliferation during root and shoot meristem development (PubMed:25564655). {ECO:0000269|PubMed:16778018, ECO:0000269|PubMed:18451323, ECO:0000269|PubMed:23095885, ECO:0000269|PubMed:24821957, ECO:0000269|PubMed:24824485, ECO:0000269|PubMed:25564655, ECO:0000269|PubMed:25752924}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
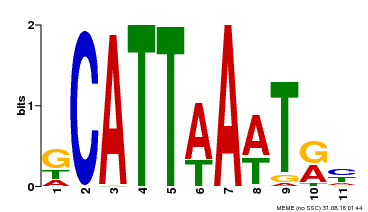
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_004515249.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004515249.1 | 0.0 | homeobox-leucine zipper protein HDG11 | ||||
| Swissprot | Q9FX31 | 0.0 | HDG11_ARATH; Homeobox-leucine zipper protein HDG11 | ||||
| TrEMBL | A0A1S2Z545 | 0.0 | A0A1S2Z545_CICAR; homeobox-leucine zipper protein HDG11 | ||||
| STRING | XP_004515249.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1026 | 33 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



