 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008218241.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 677aa MW: 76337.7 Da PI: 6.5837 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 48.5 | 2.2e-15 | 149 | 208 | 2 | 66 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkike 66
W+++e laL+++r+ m++++ + We+vs+k++e gf+rs+++Ckek+e+ ++++ +i+
XP_008218241.1 149 WSNDELLALLRIRSTMDNWFPEF-----TWEHVSRKLAELGFKRSAEKCKEKFEEESRYFNNINF 208
********************998.....9*******************************99875 PP
| |||||||
| 2 | trihelix | 97.4 | 1.3e-30 | 505 | 594 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgk.....lkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+++evlaLi++r ++ ++ ++++ k+plWe++s+ m e+g++rs+k+Ckekwen+nk+++k+k+ +kkr s +s+tcpyf+ql
XP_008218241.1 505 RWPRDEVLALINLRCSLFNNGSADQdkngvVKAPLWERISQGMLEKGYKRSAKRCKEKWENINKYFRKTKDVNKKR-SLDSRTCPYFHQLS 594
8********************9865556669********************************************8.9999********95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 6.5 | 145 | 202 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 4.9E-10 | 147 | 209 | No hit | No description |
| PROSITE profile | PS50090 | 6.377 | 148 | 200 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.24 | 502 | 569 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 2.32E-24 | 504 | 574 | No hit | No description |
| Pfam | PF13837 | 1.1E-18 | 504 | 594 | No hit | No description |
| PROSITE profile | PS50090 | 6.47 | 505 | 567 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 677 aa Download sequence Send to blast |
MFDGVPAEQL HQFIAASRTS LPLPLPLPLP LPSFPAALHA SSSPPPNINT FSAVAAAAAP 60 FDPNYNNNPS HHHLHHHHQL LLQPQPQPQP QPHQHQLLNN PGVQLHRQSA TPKNHEEKKE 120 SNLVSINNNL EIERERSSSI SQVPISDPWS NDELLALLRI RSTMDNWFPE FTWEHVSRKL 180 AELGFKRSAE KCKEKFEEES RYFNNINFTK NYRFLSDLEE LCHGGDDDQN PDQAAAGAEN 240 KNQQKVEKPS NNEGDEDNRC QILDEDSTRN ETVVGSKEFD DQDKEKEVVE RTKSNVVRKR 300 KRKRRFEMLK GFCEDIVNRM MAQQEEMHSK LLEDMVKRSE EKLAREEAWK KQEMDRMNKE 360 LEIMAHEQAI AGDRQATIIK FLKKFASSSS SSTSSEPSPD HDRRTNSSSL INQARNPNLP 420 TCSQEKEPTS STIIQKPGTS SHTPNNPSTP ISLTETTAPQ SPSSSTLAPT PTIPKVPIPP 480 ENPSSDHLNT QNPTSNDDKQ DLGKRWPRDE VLALINLRCS LFNNGSADQD KNGVVKAPLW 540 ERISQGMLEK GYKRSAKRCK EKWENINKYF RKTKDVNKKR SLDSRTCPYF HQLSTLYNQG 600 ILVPPSDHIQ GPDHQTSSAS PENQSLASPV VAQTGLDSSD QGRSSADDDL SKHNIIGEGE 660 KNNTVQPVPA AAFDFEF |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 298 | 304 | KRKRKRR |
| 2 | 299 | 304 | RKRKRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
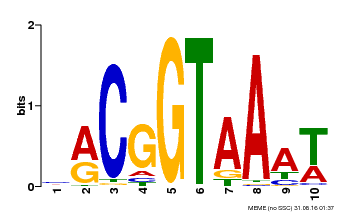
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008218241.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008218241.1 | 0.0 | PREDICTED: trihelix transcription factor GTL2 | ||||
| Swissprot | Q8H181 | 1e-106 | GTL2_ARATH; Trihelix transcription factor GTL2 | ||||
| TrEMBL | M5W7S8 | 0.0 | M5W7S8_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008218241.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5043 | 33 | 55 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 1e-103 | Trihelix family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



