 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008220039.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1024aa MW: 112295 Da PI: 5.1272 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.5 | 1.5e-18 | 37 | 83 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde+l +av+++ g++Wk+Ia+++ Rt+ qc +rwqk+l
XP_008220039.1 37 KGQWTPEEDEILRRAVQRFKGKNWKKIAECFK-DRTDVQCLHRWQKVL 83
799****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 66.5 | 4.9e-21 | 89 | 135 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEde+++++vk++G++ W+tIa++++ gR +kqc++rw+++l
XP_008220039.1 89 KGPWSKEEDEIIIELVKKYGPKKWSTIAQHLP-GRIGKQCRERWHNHL 135
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 47.1 | 5.5e-15 | 141 | 183 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT++E++ l++a++++G++ W+ + ++ gRt++ +k++w+
XP_008220039.1 141 KEAWTQDEELALIRAHQMYGNK-WAELTKFLP-GRTDNAIKNHWN 183
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.177 | 32 | 83 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.2E-15 | 36 | 85 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.7E-16 | 37 | 83 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 6.03E-16 | 38 | 93 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-24 | 39 | 95 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.07E-14 | 40 | 83 | No hit | No description |
| PROSITE profile | PS51294 | 34.567 | 84 | 139 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.05E-31 | 86 | 182 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.2E-20 | 88 | 137 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.7E-19 | 89 | 135 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.97E-18 | 91 | 135 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-28 | 96 | 139 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.147 | 140 | 190 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.1E-14 | 140 | 188 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.8E-21 | 140 | 190 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.1E-13 | 141 | 184 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.28E-11 | 143 | 183 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1024 aa Download sequence Send to blast |
MEGDRTNSTP SEGLGDSIQK VRALHGRTSG PTRRSTKGQW TPEEDEILRR AVQRFKGKNW 60 KKIAECFKDR TDVQCLHRWQ KVLNPELVKG PWSKEEDEII IELVKKYGPK KWSTIAQHLP 120 GRIGKQCRER WHNHLNPGIN KEAWTQDEEL ALIRAHQMYG NKWAELTKFL PGRTDNAIKN 180 HWNSSVKKKL DSYLKSGLLT QFQGLPHVGH QNQSVLSSSS RMQSSGDDSG AKAAEGEEIS 240 ECSQDSTVAG CFLSATEMTN VVPHPREEFQ INEVSRLGND PSCSPASCSE PYYPSVGDAT 300 FSIPEIPPEM VCSKFIEQNF SHEAGASMSG DFQFNLHELP INSSLECGQE SSRMHTHCVG 360 CNESHEGANA PFQTSTSMGN MAVGFIKSEH MLISDDECCR VLFSDAMNGG CFSSGDFTNG 420 ANMVDLGACT DSVLLQPSNL QISETGRTSA SQVYHPLSSD VTGTSCSQVV SAHEGPLIYA 480 GEPSHLFRVQ EQEFVTNSND GFIYANDSAS NDTGMQEQSD LVKDPSKLVP VNTFDSGLDL 540 QNCPVDVRSD EQTEQQDGGA LCYEPPRFPS LDIPFFSCDL VQSGNDMQQX NCLTPYRLWD 600 SPSRESSPDA VLKSAAKTFT GTPSILKKRH RDLLSPLSPL SDRRIDKRLG TDLTSSLARD 660 FSRLDVMFED SEEKTTLLSP SSNKNRNSDS PSEDKENKGT CESRIEKGTD STALSDDGIA 720 HKDFDNGESQ EKTKQFQGIA DIDAKNKVDV VPTSQIAQQT SGVLVEHNAN DLLLCSPVGC 780 KAEKALGTST KTPRSQFRKS FEATNPGVPS KSFSARQCAS VKSPTICVKK HESYSLVDTC 840 VQSDSLSAPP ETTGDNAGND ISIENIFGDT PFKRSIESPS AWKSPWFINS FVPGPRVDTE 900 ISIEDIGFFM SPGDRSYDAI GLMKQISEQT AAAYANAQEV LGNETPETLF RERRKNQALV 960 DPENNHGPPN QPGSSSLSAA NVLVERRTLD FSECGTPGKG IENAKSSNAK TFSSPSSYLL 1020 KGCR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-69 | 37 | 190 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-69 | 37 | 190 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00466 | DAP | Transfer from AT4G32730 | Download |
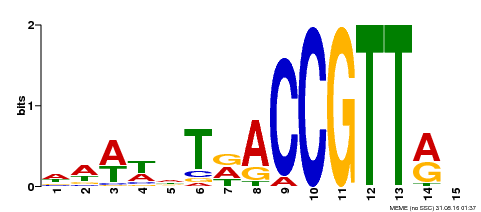
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008220039.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008220039.1 | 0.0 | PREDICTED: LOW QUALITY PROTEIN: myb-related protein 3R-1-like | ||||
| TrEMBL | A0A251RDM5 | 0.0 | A0A251RDM5_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008220039.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3727 | 33 | 62 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32730.2 | 0.0 | Homeodomain-like protein | ||||



