 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008223308.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 898aa MW: 101632 Da PI: 7.2 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 163.6 | 3.4e-51 | 26 | 142 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
+e k+rwl+++ei+aiL n++ +++ ++ + p+sg+++L++rk++r+frkDG++wkkk dgktv+E+he+LKvg+ e +++yYah+e+ ptf r
XP_008223308.1 26 EEaKSRWLRPNEIHAILYNHKYFTIYVKPVNLPQSGTIVLFDRKMLRNFRKDGHNWKKKNDGKTVKEAHEHLKVGNEERIHVYYAHGEDSPTFVR 120
4559******************************************************************************************* PP
CG-1 97 rcywlLeeelekivlvhylevk 118
rcywlL+++le+ivlvhy+e++
XP_008223308.1 121 RCYWLLDKSLEHIVLVHYRETQ 142
*******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 76.289 | 21 | 147 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.4E-76 | 24 | 142 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.5E-45 | 27 | 140 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.31E-13 | 352 | 439 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 5.36E-14 | 512 | 646 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.7E-16 | 536 | 650 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 6.53E-17 | 549 | 649 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 5.7E-7 | 549 | 616 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.3 | 554 | 658 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.621 | 587 | 619 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 9.2E-6 | 587 | 616 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1600 | 626 | 656 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.43E-5 | 726 | 801 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.73 | 735 | 761 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 26 | 750 | 772 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.053 | 751 | 780 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.022 | 773 | 795 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.164 | 774 | 798 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 8.0E-4 | 775 | 795 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 46 | 853 | 875 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.199 | 854 | 883 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 898 aa Download sequence Send to blast |
MEKQLEGSEI HGFHTMQDLD VGTIMEEAKS RWLRPNEIHA ILYNHKYFTI YVKPVNLPQS 60 GTIVLFDRKM LRNFRKDGHN WKKKNDGKTV KEAHEHLKVG NEERIHVYYA HGEDSPTFVR 120 RCYWLLDKSL EHIVLVHYRE TQELQGSPVT PVNSNNSSSV SDPSAPWLLS EELDSGANKS 180 YCAGENELSE PGDGLTVKNH EKRLHDINTL EWEELLITND SKGDIVSCYD QQNQVVGNGF 240 ISGGASVISA EMSAFDNLTN PTSRSDNVQF NLLDSPYVPT VEKTTYDSLD VLVNDGLHSQ 300 DSFGRWINQV MADPPGSVED PALESSSLAA QNSFASPSAD HLQSSVPHQI FNITDLSPAW 360 AFSNEKTKIL ITGFFHQEYL HLAKSDLLCI CGDVCLRAEI VQAGVYRCFV PPHLPRVVNL 420 FMSIDGHKPI SLVLNFEYRA PVLSDPIISS EENKWEEFQA QMRLAYLLFS SSKNLNIVSN 480 KVLPNALKEA KKFSHRTSHI SNSWAYLMKA VEDNKTPLPL AKDGLFELIL KNRLKDWLLE 540 KVVASSTTKE YDAYGQGVIH LCAILEYTWA VRLFSWSGLS LDFRDRRGWT ALHWAAYCGR 600 EKMVAVLLSA GAKPNLVTDP SSENPGGCTA ADLAAMKGYD GLAAYLSEKA LVEQFKDMSM 660 AGNASGSLQT SSNYAGNSEN LSEDEIHLKD TLAAYRTAAD AAARIQAAFR ENSLKLKAKA 720 VQYSTPEAEA RGIIAALKIQ HAFRNYDTRK KIKAAARIQY RFRTWKMRQE FLSLRRQAIK 780 IQAAFRGFQV RRQYRKVLWS VGVLEKAVLR WRFKRRGLRG LNVAPVEVDV DQKQESDTEE 840 DFYRASRKQA EERIERSVVR VQAMFRSKKA QEEYSRMKLT HIEAKLEFEE LLNPDLDS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
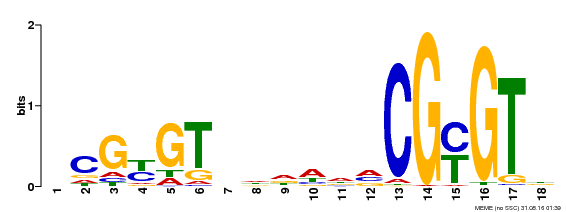
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008223308.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC208139 | 1e-38 | AC208139.1 Populus trichocarpa clone JGIACSB13-D20, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008223308.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A251QWG4 | 0.0 | A0A251QWG4_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008223308.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4288 | 34 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



