 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008224267.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 794aa MW: 87204.4 Da PI: 6.0965 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 49.8 | 7e-16 | 497 | 535 | 2 | 40 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
++k+CnCkkskClk+YCeCfaag++C e C+C++C+Nk
XP_008224267.1 497 SCKRCNCKKSKCLKLYCECFAAGVYCIEPCSCQECFNKP 535
689**********************************96 PP
| |||||||
| 2 | TCR | 50.5 | 4.1e-16 | 582 | 620 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
XP_008224267.1 582 RHKRGCNCKKSSCLKKYCECYQGGVGCSISCRCEGCKNA 620
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 2.5E-17 | 496 | 537 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.23 | 497 | 622 | IPR005172 | CRC domain |
| Pfam | PF03638 | 1.6E-11 | 499 | 534 | IPR005172 | CRC domain |
| SMART | SM01114 | 4.4E-17 | 582 | 623 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 1.3E-11 | 584 | 620 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 794 aa Download sequence Send to blast |
MDTPERNQIG TPKAKFEDSP VFNYINSLSP IKPVKSVHIT QTFSSLSFAS LPSVFTSPHV 60 SAHKESRFLR RHSTSDPSKP ESSLESGNKV SANEDAAQLY INSEELREDC VPGVSTGEDS 120 VEPSSEHSKF VIELPRNLKY DCGSPDCRPT TRCGTEAHCE LEVADLSAPL VPYVQKTSEE 180 GSSNDEAHLQ IICQTLQRKE GTGCDWESLI SDAADLLIFD SPNGTEAFKG LMQNSLDPAT 240 RFCTSLAPQL TQNDVNDEQN MQVLDIVGSG RQLETEDPSS QYGEASQLER TEQMEDHLNN 300 CMVSSQSEKE DNKVETPMQF NCKPAVLNLQ RGLRRRCLDF EMAGARRKSL DNVSNSSSNM 360 LSQSDEKITT NDKQLVPMKP GGESSRCILP GIGLHLNALA KTSKDYKIIK CESLAYGRQL 420 SLPNSTADIH SPTGGQGPGH ESFSSASSER DMDGTENGVQ LAHDASQEPA FLANEEFNQN 480 SPKKKRRRFE QAGETESCKR CNCKKSKCLK LYCECFAAGV YCIEPCSCQE CFNKPIHEDT 540 VLATRKQIES RNPLAFAPKV IRSSDSVPEL GEESSKTPAS ARHKRGCNCK KSSCLKKYCE 600 CYQGGVGCSI SCRCEGCKNA FGRKDGSFIG TEAELDEEEA EACEKSVAEK HQQKIEIQKN 660 EEQRLDSALP TTPLRLSRQM VSLPFSSKNK PPRSSVFSIG GSSSGLYTSQ KLGKPNILRP 720 ESKFERHSQT VPEDEMPEIL QGDGSPSTGV KTASPNSKRV CPPNSDFGPS PGRRTGRKLI 780 LQSIPSFPSL TPQH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 4e-17 | 499 | 619 | 12 | 120 | Protein lin-54 homolog |
| 5fd3_B | 4e-17 | 499 | 619 | 12 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 482 | 487 | KKKRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
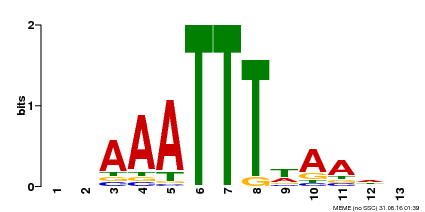
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008224267.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008224267.1 | 0.0 | PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 | ||||
| TrEMBL | A0A251QRB3 | 0.0 | A0A251QRB3_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008224266.1 | 0.0 | (Prunus mume) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 1e-144 | TESMIN/TSO1-like CXC 2 | ||||



