 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008229656.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 895aa MW: 102695 Da PI: 6.6621 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 84.8 | 1.3e-26 | 136 | 238 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk............tekerrtraetrtgCkaklkvkkekdgkwevtklel 83
+fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k+ e++ +r+ ++t+Cka+++vk++ dgkw+++++++
XP_008229656.1 136 SFYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYDKSynrprarqnkqdPENATGRRSCSKTDCKASMHVKRRPDGKWVIHNFVK 230
5******************************************99999999988776554555559999************************** PP
FAR1 84 eHnHelap 91
eHnHel p
XP_008229656.1 231 EHNHELLP 238
*****975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 1.9E-24 | 136 | 238 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 1.5E-33 | 336 | 428 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 8.889 | 616 | 652 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 1.9E-6 | 627 | 654 | IPR006564 | Zinc finger, PMZ-type |
| Pfam | PF04434 | 2.1E-4 | 628 | 650 | IPR007527 | Zinc finger, SWIM-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 895 aa Download sequence Send to blast |
MLEEEDVIIF LTLTRLDNLV GSVSYIVLRM SKMIFSDNNQ LKLNNLTMDI DLRLPSGEPD 60 KEDEEPHGID NMLEHEEKLQ NGDIENGNIV DVRDEVHAED GGDLNSPTAD MVVFKEDTNL 120 EPLFGMEFAS HGEAYSFYQE YARSMGFNTA IQNSRRSKTS REFIDAKFAC SRYGTKREYD 180 KSYNRPRARQ NKQDPENATG RRSCSKTDCK ASMHVKRRPD GKWVIHNFVK EHNHELLPAQ 240 AVSEQTRKMY AAMARQFAEY KNVVGLKNDP KNPFDKGRNL ALEAGDLKIL LDFFTQMQNM 300 NSNFFYAIDL GEDQRLKSLF WVDAKSRHDY INFSDVVSFD TTYIRNKYKM PLVLFVGVNQ 360 HYQFVLLGCA LVSDESTTTF SWLMQTWLKA MGGQAPKVII TDHDKSIKSV ISEVFPNAYH 420 CFCLWHILGK VSENLGHVIK RHENFMAKFE KCIHRSSTNE EFEKRWWKIL EKFELKDDEW 480 TQSLYEDRKQ WVPTYMRDVC LAGMSVVQRS ESVNSFFDKY VHKKTTVQEF LKQYEAILQD 540 RYEEEAKADS DTWNKQPTLR SPSPLEKSVS GVYTHAVFKK FQVEVLGAVA CHPKREGQDE 600 TTITFRVQDF EKNQDFIVTW NEMKTEVSCL CCLFEYKGYL CRHALIVLQI CGLSAIPVQY 660 ILKRWTKDVK NRHLVGEESD HGLSRVQKFN DLCQRAMKVI EEGSLSQESY SVACRALEEA 720 FGNCVSVNNS SKSLIEAGTS SVTHGLLCIE DDSQNRSMGK TNKKKNPTKK RKVNSEPDVM 780 TVGAQDSLQQ MDKLNPRAVT LDGYYGAQQS VQGMVQLNLM APTRDNYYGN QQTIQGLGQL 840 NSIAPSHDGY YSAQQSMHGL GQMDFFRTPG GFTYGMRDDP NVRTAPLHDD ASRHA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
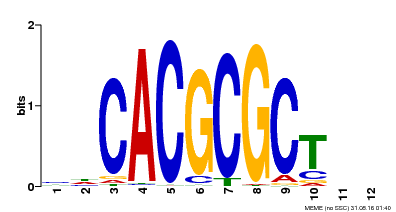
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008229656.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008229656.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A251Q297 | 0.0 | A0A251Q297_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008229656.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF40 | 30 | 530 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



