 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008230943.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 467aa MW: 52555.9 Da PI: 7.3868 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.9 | 3.5e-16 | 253 | 299 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT++Ed llv++v ++G + W+ Ia+++ gR +kqc++rw+++l
XP_008230943.1 253 KGQWTPQEDRLLVQLVGRFGIKKWSQIAKMLS-GRVGKQCRERWHNHL 299
799*****************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 57 | 4.6e-18 | 306 | 347 | 2 | 45 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ W++eEd++l++a+kq G++ W+ Ia++++ gRt++ +k++w+
XP_008230943.1 306 DMWSEEEDKILIEAHKQIGNK-WAEIAKMLP-GRTENTIKNHWN 347
67*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 28.409 | 248 | 303 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.11E-32 | 250 | 346 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.1E-14 | 252 | 301 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-23 | 254 | 303 | IPR009057 | Homeodomain-like |
| Pfam | PF13921 | 1.8E-17 | 256 | 313 | No hit | No description |
| CDD | cd00167 | 4.77E-14 | 256 | 299 | No hit | No description |
| PROSITE profile | PS51294 | 18.89 | 304 | 354 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.4E-15 | 304 | 352 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-21 | 304 | 352 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.12E-13 | 308 | 350 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010439 | Biological Process | regulation of glucosinolate biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1904095 | Biological Process | negative regulation of endosperm development | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 467 aa Download sequence Send to blast |
MELETNLKEL DNFPFHSSLF SDSSTPMKLK PEHPSNTNNG INSLRPQLSS SSPPPPNCKP 60 LFSSLVPHFD PHHQHLHLHH HHHHQPLHGS HLNLNSHSHD DFKTSSDHIN HFSTIEGSSS 120 NPFSGIYYDT NPHDFVAVPL APDGIDFHGA RGSYWGAHHH HHLDGQEIPD DQAQMGPQSC 180 IQLPPLINFG EFGSSSAMAI SLPSDEVTCN SSSSTEIEFC KRLSADDQKK TKRLCMRRPT 240 SSKVLQKKPS IIKGQWTPQE DRLLVQLVGR FGIKKWSQIA KMLSGRVGKQ CRERWHNHLR 300 PDIRKDMWSE EEDKILIEAH KQIGNKWAEI AKMLPGRTEN TIKNHWNATK RKQNSKKKTN 360 KDPNNNINPP ESSLLQVYIR SVVSSSSAFS SSSQMGGSVS NLFNFESYGF GGSFLVDQVT 420 GVEEYRGSSM EAEMPLALEV KSLMQGPDDD HVKKEMDLLE MICQGKL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 5e-45 | 249 | 355 | 3 | 110 | B-MYB |
| 1mse_C | 3e-45 | 251 | 353 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 3e-45 | 251 | 353 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00385 | DAP | Transfer from AT3G27785 | Download |
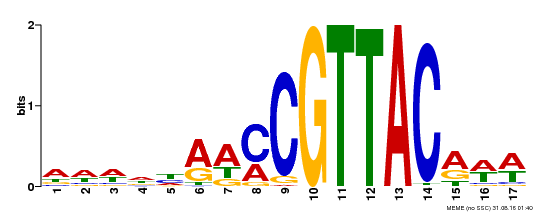
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008230943.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008230943.1 | 0.0 | PREDICTED: transcription factor MYB98 | ||||
| TrEMBL | M5X7L2 | 0.0 | M5X7L2_PRUPE; Uncharacterized protein (Fragment) | ||||
| STRING | XP_008230943.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF17095 | 4 | 8 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18770.1 | 8e-60 | myb domain protein 98 | ||||



