 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008233952.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 367aa MW: 41599.7 Da PI: 6.8065 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 36 | 1.5e-11 | 80 | 119 | 3 | 42 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeN 42
++k++rr+++NReAAr+sR RKka++++Le+ +L +
XP_008233952.1 80 SDKVQRRLEQNREAARKSRMRKKAYVQQLETSRLKLAQLE 119
79****************************8544443333 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 3.0E-10 | 75 | 128 | No hit | No description |
| SMART | SM00338 | 1.8E-5 | 78 | 138 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.4E-8 | 80 | 120 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.289 | 80 | 122 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.0E-8 | 82 | 124 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 85 | 100 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 1.9E-26 | 171 | 244 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 367 aa Download sequence Send to blast |
MLQERMSYPS TQFATSRQMG IYEPFRQVGV WKETFRGDHS SNTGASSILE VDAGIETKIG 60 YVSHESLAPS GNDQEPNRSS DKVQRRLEQN REAARKSRMR KKAYVQQLET SRLKLAQLEQ 120 ELDRARKQGA YVASSLGTSH MGYNGALNSG ITTFEMEYGH WVEEQHRQNV ELRNALQDHG 180 TDIELLRILV ESGLSHYANL FRMKADAARA DAFYLLSGIW RTSVERHFHW IGGFRPSELL 240 NIVLPQLQPL DDPQFVDFCN LRQSSQQAED ALTQGMEKLQ QNLALTIAGD QISGESYGSQ 300 MATALEKLEA LETFVGQADH LRQQTLQQMS RILTTHQAAR GLLALGEYFH QLRALSSLWT 360 ARPRELT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
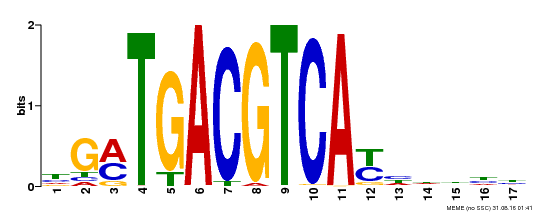
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1 (By similarity). {ECO:0000250, ECO:0000269|PubMed:12953119}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00247 | DAP | Transfer from AT1G77920 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008233952.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008233952.1 | 0.0 | PREDICTED: transcription factor TGA3 isoform X3 | ||||
| Swissprot | Q93ZE2 | 1e-152 | TGA7_ARATH; Transcription factor TGA7 | ||||
| TrEMBL | A0A314UVR0 | 0.0 | A0A314UVR0_PRUYE; Transcription factor TGA3 isoform X3 | ||||
| STRING | XP_008233950.1 | 0.0 | (Prunus mume) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G77920.1 | 1e-154 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



