 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008235184.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 543aa MW: 60313.2 Da PI: 4.8504 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.8 | 1.8e-16 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+grWT+eEd++l+++++ G g+W++ ++ g+ R++k+c++rw +yl
XP_008235184.1 14 KGRWTAEEDQILINYIQTNGEGSWRSLPKNAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 48.8 | 1.6e-15 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ +++E+++++++++ lG++ W++Ia+ ++ gRt++++k++w+++l
XP_008235184.1 67 RGNISAQEEDIIIKLHASLGNR-WSLIASQLP-GRTDNEIKNYWNSHL 112
7899******************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 8.4E-22 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.678 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.03E-27 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.8E-12 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.4E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.64E-9 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 24.903 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-24 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.2E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.4E-14 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.13E-10 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009813 | Biological Process | flavonoid biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 543 aa Download sequence Send to blast |
MGRAPCCDKI GLKKGRWTAE EDQILINYIQ TNGEGSWRSL PKNAGLLRCG KSCRLRWINY 60 LRADLKRGNI SAQEEDIIIK LHASLGNRWS LIASQLPGRT DNEIKNYWNS HLSRKIDTFR 120 RPTTTTSDQM SSLPAAASNN IPSKRRGGRT SRWAMKKSKT YTTTNSTNYT QRHNKGQKEI 180 TNIAAADDEA IALEKKTSLP GPNDNIIDTM HRDYMVLMTD PAADHHDHHP HHMDDCRVDN 240 LVNHDHDHQQ QEEAGGLAIP VVLMSSTTTT TTTEDEDEKE EVVVEVEVEK KETRDHDGLC 300 PVNIDQCQKE SQEMLLGPHP HNDENKDDDL DESGGIEFDG GLLGTFNELM DNVELLQKDP 360 NSNGVLSDHQ HDHAMGVTHE VYQVETTTTA TATTCGHLSS SSNGQLCFSS IMSMTATSSS 420 SASASAAAGS YYFDMEDGQA AAANGNDRDH NNHILWDWES VVEAGHELWD HDDDKENMLS 480 WLWEEGTCSS STTSNTTAAA AAAANTIDRH LNWEGDTTTY DTSMMRKAVD ADKQNAMVAW 540 LLS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 7e-24 | 12 | 118 | 5 | 110 | B-MYB |
| 1h8a_C | 8e-24 | 12 | 116 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00627 | PBM | Transfer from PK06182.1 | Download |
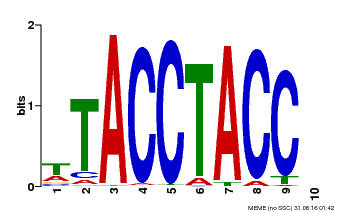
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008235184.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU938682 | 0.0 | GU938682.1 Prunus avium R2R3-MYB transcription factor (MYBF1) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008235184.1 | 0.0 | PREDICTED: transcription factor MYB29 | ||||
| TrEMBL | A0A251N776 | 0.0 | A0A251N776_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008235184.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47460.1 | 3e-75 | myb domain protein 12 | ||||



