 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008236589.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 429aa MW: 46634.4 Da PI: 10.4296 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 45 | 2.4e-14 | 351 | 401 | 5 | 55 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkke 55
+r+rr++kNRe+A rsR+RK+a++ eLe +v++L++ N++L+ + e+ ++
XP_008236589.1 351 RRQRRMIKNRESAARSRARKQAYTLELEAEVAKLKEMNEELQRKQTEIMEM 401
79************************************9998877777666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.3E-11 | 347 | 412 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.76 | 349 | 400 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.7E-12 | 351 | 401 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 5.08E-25 | 351 | 405 | No hit | No description |
| SuperFamily | SSF57959 | 3.8E-10 | 351 | 399 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 4.3E-14 | 351 | 400 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 354 | 369 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 429 aa Download sequence Send to blast |
MGSNFNFKNF GDVPPGEGNG GKAAGNFMLA RQSSVYSLTF DEFQNTIGGL GKDFGSMNMD 60 ELLKNIWTAE ETQGVTSTSG AGEGSAPGGN LQRQGSLTLP RTLSQKTVDE VWKDLIRETS 120 DAKYNTVAMG SNLPQRQQTL GEMTLEEFLV RAGVVREDVQ PVVRPNNSGF YGELYRPNNH 180 NGIAPGFQQP SRTNGLLGNR VADNNNSVLN QSPNLALNVG GVRSSQQQTQ QLPPQQQPLF 240 PKPTNVAFAP SMHLTNNAQL TSPRTRGPMT GVVEPSMNTV FTQGGGFPGA GIGMTGLGTG 300 GGAVAARSPT NQISPDVIAK SSGDTSSLSP VPYMFSRGRK SNGALEKVVE RRQRRMIKNR 360 ESAARSRARK QAYTLELEAE VAKLKEMNEE LQRKQTEIME MQKDQILETV KRQRGGKRQC 420 LRRTLTGPW |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. {ECO:0000269|PubMed:11884679, ECO:0000269|PubMed:15361142}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
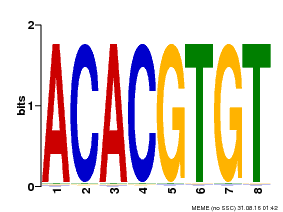
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008236589.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA). {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:16284313}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008236589.1 | 0.0 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 | ||||
| Swissprot | Q9M7Q3 | 1e-113 | AI5L6_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 6 | ||||
| TrEMBL | A0A251MWZ9 | 0.0 | A0A251MWZ9_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008236589.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1962 | 34 | 81 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G34000.2 | 1e-100 | abscisic acid responsive elements-binding factor 3 | ||||



