 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008237396.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 577aa MW: 65456.8 Da PI: 4.7388 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 167.9 | 3.5e-52 | 18 | 144 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevls 94
+pGfrFhPtdeelvv+yLk+k+ +k+l+l +vi+e+d+yk++P++Lp + +k+++++w+fFs+rd+ky++g r+nrat++gyWkatgkd+++++
XP_008237396.1 18 APGFRFHPTDEELVVYYLKRKICKKRLKL-NVIAETDVYKWDPEELPglSLLKTGDRQWFFFSPRDRKYPNGGRSNRATRHGYWKATGKDRNITC 111
69***************************.99***************644788899*************************************** PP
NAM 95 kkgelvglkktLvfykgrapkgektdWvmheyrl 128
++ vglkktLv+ykgrap+ge+tdWvmhey+l
XP_008237396.1 112 -YSRSVGLKKTLVYYKGRAPSGERTDWVMHEYTL 144
.9999***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.84E-58 | 12 | 167 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 54.183 | 17 | 167 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.6E-27 | 19 | 144 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0070301 | Biological Process | cellular response to hydrogen peroxide | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 577 aa Download sequence Send to blast |
MGSEDDPEML FRNGRFCAPG FRFHPTDEEL VVYYLKRKIC KKRLKLNVIA ETDVYKWDPE 60 ELPGLSLLKT GDRQWFFFSP RDRKYPNGGR SNRATRHGYW KATGKDRNIT CYSRSVGLKK 120 TLVYYKGRAP SGERTDWVMH EYTLDEEELK RCRNVQEYYA LYKVYKKSGP GPKNGEQYGA 180 PFREEEWADD ELPVINNPAD RQIPVKQSVD VISVDPVKVN GEVHSALSDI EEFMKQIADE 240 AVLELPQMNG YAYTIPQAVS EEETQSTVVD LYSREVVCPE PNTVFNPSHH QGNLQASFDF 300 TQSDTSQIQR YEASEVTTSA PEIHEQGPPI LREEDFLEMD DLLGPEPTIS NIENPVDNLQ 360 FEGIDGLSEF DLYHDAAMFF HDMEPFDQGT VSHQQYMNSL GNNIVDQFEY QLQPNPPAVN 420 QVNHQLNPES TQMNNQLWTH TERAEPNQGC LSYSTSGVVY EPSNFASQAN QNQSGNEAAG 480 GPSQFSSALW AFVESIPTTP ASASENALVN RAFERMSSFS RLRINSVSAN VTAGSSSEAR 540 RAGRKRGFFF LPVVVALCAI FWVLMATLRP WWRCLSA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 5e-47 | 19 | 144 | 17 | 140 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 35 | 44 | KRKICKKRLK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP). Transcriptional activator that acts as positive regulator of AOX1A during mitochondrial dysfunction. Binds directly to AOX1A promoter. Mediates mitochondrial retrograde signaling. {ECO:0000269|PubMed:24045017}. | |||||
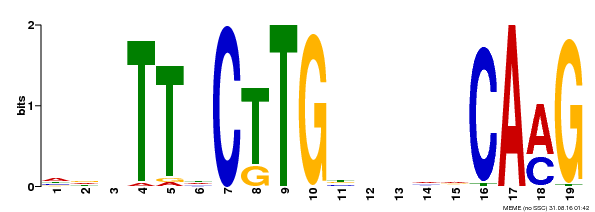
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00179 | DAP | Transfer from AT1G34190 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008237396.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold and drought stresses. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008237396.1 | 0.0 | PREDICTED: NAC domain-containing protein 17 | ||||
| Swissprot | Q9XIC5 | 1e-170 | NAC17_ARATH; NAC domain-containing protein 17 | ||||
| TrEMBL | M5W6F5 | 0.0 | M5W6F5_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008237396.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6131 | 34 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34190.1 | 1e-156 | NAC domain containing protein 17 | ||||



