 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008237582.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 591aa MW: 66570.5 Da PI: 6.6406 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 91 | 1.2e-28 | 66 | 150 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW++qe+laL+++r++m+ ++r+++lk+plWe+vs+k+ e g+ rs+k+Ckek+en+ k+++++keg++++ ++ +t+++fd+lea
XP_008237582.1 66 RWPRQETLALLKIRSQMDAAFRDSSLKAPLWEDVSRKLGELGYYRSAKKCKEKFENVYKYHRRTKEGRSGK--QEGKTYRFFDELEA 150
8********************************************************************96..45557******985 PP
| |||||||
| 2 | trihelix | 100.1 | 1.9e-31 | 413 | 497 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW++ ev aLi++r+ ++ r++++ k+ lWee+s+ mr+ g++rs+k+Ckekwen+nk++kk+ke++k r +e+s+tcpyf+qle
XP_008237582.1 413 RWPRVEVEALINLRTCLDVRYQEAGPKGSLWEEISAGMRQLGYNRSAKRCKEKWENINKYFKKVKESSKTR-PEDSKTCPYFNQLE 497
8********************************************************************97.99***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.001 | 63 | 125 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 1.0E-18 | 65 | 150 | No hit | No description |
| CDD | cd12203 | 1.10E-23 | 65 | 130 | No hit | No description |
| PROSITE profile | PS50090 | 6.818 | 65 | 123 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.0072 | 410 | 472 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 2.7E-21 | 412 | 498 | No hit | No description |
| CDD | cd12203 | 1.59E-27 | 413 | 477 | No hit | No description |
| PROSITE profile | PS50090 | 6.957 | 413 | 470 | IPR017877 | Myb-like domain |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 591 aa Download sequence Send to blast |
MPAVDVPAPD SSSTMLAAGS SSGEGLAGVV REGSHETTAV LSNSVQEDKD GRVDDLGDRN 60 LYGGNRWPRQ ETLALLKIRS QMDAAFRDSS LKAPLWEDVS RKLGELGYYR SAKKCKEKFE 120 NVYKYHRRTK EGRSGKQEGK TYRFFDELEA FDQQNNHPSV PPKPQVLLWP NNNHHHPNNP 180 TVVSHGITSV VPLSSTPNMM ATTNGSLNGS HISPPPINSL PPQVTKPVNL SHQNLNGFKP 240 SLATSNLFSS STSSTASDEE FQVQQSGKKK RKWKYFFRRL TKEVLEKQEK LQEKFLEAIA 300 KCEHQRTVRE EAWRMQEMAR LDKEHQILAQ ERSSAAAKDA AVIEFLQKVS GQQNVTNNIQ 360 AIEVNRPIPS RPQPLPVLMA PPPPAPVAVI TTSFEVPRLD KGDNSTIPPG STRWPRVEVE 420 ALINLRTCLD VRYQEAGPKG SLWEEISAGM RQLGYNRSAK RCKEKWENIN KYFKKVKESS 480 KTRPEDSKTC PYFNQLEDLY RKKNMNNSSH VGYVEKPLPQ AILQPLIKVQ PDQPVVEAIE 540 GENGDKNEED EDGESTEEED HHLDQSNDDD QDEQMGIVTN YKESSMEMVE S |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00244 | DAP | Transfer from AT1G76890 | Download |
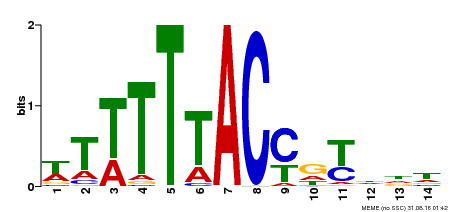
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008237582.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008237582.1 | 0.0 | PREDICTED: trihelix transcription factor GT-2-like | ||||
| Swissprot | Q39117 | 1e-135 | TGT2_ARATH; Trihelix transcription factor GT-2 | ||||
| TrEMBL | M5VKC5 | 0.0 | M5VKC5_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008237582.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF373 | 34 | 181 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76890.2 | 1e-117 | Trihelix family protein | ||||



