 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_008237894.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 340aa MW: 37503.4 Da PI: 4.4734 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 31.1 | 5e-10 | 156 | 197 | 4 | 45 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaL 45
+k+ rr +N+ AA rsR+RKk+++ Le k+k Le+e ++L
XP_008237894.1 156 SKKRRRQLRNKDAAVRSRERKKMYVRDLEMKSKYLEGECRRL 197
6999**********************************9866 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 4.8E-4 | 152 | 217 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 9.2E-11 | 152 | 212 | No hit | No description |
| PROSITE profile | PS50217 | 9.473 | 155 | 197 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.19E-10 | 157 | 212 | No hit | No description |
| Pfam | PF00170 | 3.7E-8 | 157 | 197 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14704 | 2.94E-15 | 158 | 209 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 160 | 175 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0030968 | Biological Process | endoplasmic reticulum unfolded protein response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 340 aa Download sequence Send to blast |
MELEGDVGFL DSENDIIGEI DWDFLFDGSN ILEDVLELEN PVVTATGSSS PSPSPSSTVE 60 DAPSNSSPDW IGEIETILMK DDDVNGNQVV PDSAEPTNAE FYEKFLADIL VDSPSSDAGS 120 NASADSVKEK LDRSPLNDDD DDDDDTADAD ADDPISKKRR RQLRNKDAAV RSRERKKMYV 180 RDLEMKSKYL EGECRRLGRL LQCCYAENHA LRLGLQMNNA YGHGHGVLAT KQESAVLLLE 240 LLLLGSLLRC LDIMCLVALP LILMAGLRNP LNNVTANKDL ESVDLRPRGA ASKMFQHSVL 300 QCFSKSRRCK ASRTKMKPSR FPCLLGSYVS FALPLLMLAT |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00019 | PBM | Transfer from AT1G42990 | Download |
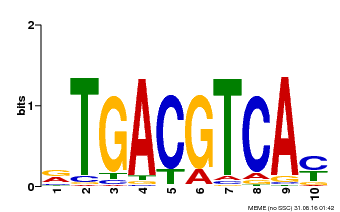
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_008237894.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT373972 | 1e-112 | KT373972.1 Pyrus pyrifolia cultivar mixueli putative bZIP transcription 60 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008237894.1 | 0.0 | PREDICTED: bZIP transcription factor 60 | ||||
| TrEMBL | M5W9Y4 | 0.0 | M5W9Y4_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008237894.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF12009 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G42990.1 | 3e-09 | basic region/leucine zipper motif 60 | ||||



