 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009104292.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 785aa MW: 85993.2 Da PI: 6.6896 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61 | 1.8e-19 | 101 | 156 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++ Le++F+++ +p++++r +L+++l+L+ rqVk+WFqNrR+++k
XP_009104292.1 101 KKRYHRHTAKQIQDLESVFKECAHPDEKQRLDLSRRLNLDPRQVKFWFQNRRTQMK 156
688999***********************************************999 PP
| |||||||
| 2 | START | 185.7 | 2.4e-58 | 301 | 520 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEE CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetle 83
la++a++elvk+a+ ep+Wv+ss ++n++e+ ++f++ + + +ea++++g+v+ ++ lve+l+d+ +W e+++ + +t+e
XP_009104292.1 301 LALAAMEELVKMAQRHEPLWVRSSdtgfVMLNKEEYDTSFSRVVGlkqdgFVSEASKETGNVIINSLALVETLMDSE-RWAEMFPsmisRTSTTE 394
6899************************77777777777755444777889**************************.*******9999****** PP
EECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE CS
START 84 vissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvd 171
issg gal+lm aelq+lsplvp R + f+R+++q+ +g+w++vdvS+ds ++ + sss+ R lpSg+l+++++ng+skvtw+eh++
XP_009104292.1 395 IISSGmggtrnGALHLMHAELQLLSPLVPvRQVSFLRFCKQHAEGVWAVVDVSIDSIREGS-SSSCRR---LPSGCLVQDMANGYSKVTWIEHTE 485
************************************************************9.777766...************************ PP
--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 172 lkgrlphwllrslvksglaegaktwvatlqrqcek 206
++++ +h l+r+l++ gla+ga++w+a+lqrqce+
XP_009104292.1 486 YDENRIHRLYRPLLSCGLAFGAQRWMAALQRQCEC 520
*********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.09E-19 | 88 | 158 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-20 | 95 | 165 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.394 | 98 | 158 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 7.9E-18 | 99 | 162 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 4.5E-17 | 101 | 156 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 6.60E-16 | 105 | 158 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 133 | 156 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 39.844 | 291 | 523 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.43E-30 | 294 | 520 | No hit | No description |
| CDD | cd08875 | 8.64E-111 | 295 | 519 | No hit | No description |
| SMART | SM00234 | 1.0E-42 | 300 | 520 | IPR002913 | START domain |
| Pfam | PF01852 | 2.7E-50 | 301 | 520 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.16E-18 | 549 | 777 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 785 aa Download sequence Send to blast |
MNFNGYLGDG SSRDRISDVP YSDNFSFSAA ATMPHSRPFS SNGLSLGLQT NGVNGEAFET 60 NVTRNKSRGG EDVESRSESD NAEAVSGDDL ETGDKPPRKK KKRYHRHTAK QIQDLESVFK 120 ECAHPDEKQR LDLSRRLNLD PRQVKFWFQN RRTQMKTQIE RHENALLRQE NDKLRAENMS 180 VREAMRNPMC SNCGGPAVLG EVSMEEQHLR IENSRLKDEL DRVCALTGKF LGRSPSGSHH 240 VPDSSLVLGV GVGSGGGFTL SSPSLPQASP RFEISNGTGL ATVNIQAPVS DFDQRSRYLD 300 LALAAMEELV KMAQRHEPLW VRSSDTGFVM LNKEEYDTSF SRVVGLKQDG FVSEASKETG 360 NVIINSLALV ETLMDSERWA EMFPSMISRT STTEIISSGM GGTRNGALHL MHAELQLLSP 420 LVPVRQVSFL RFCKQHAEGV WAVVDVSIDS IREGSSSSCR RLPSGCLVQD MANGYSKVTW 480 IEHTEYDENR IHRLYRPLLS CGLAFGAQRW MAALQRQCEC LTILMSSTVS PSRSPTPISC 540 NGRKSMLKLA KRMTDNFCGG VCASSLQKWS KLNVGNVDED VRIMTRKSVD DPGEPPGIVL 600 NAATSVWMPV SPKRLFDFLG NERLRSEWDI LSNGGPMQEM AHIAKGHDHS NSVSLLRATA 660 INANQSSMLI LQETSIDAAG AVVVYAPVDI PAMQAVMNGG DSAYVALLPS GFAILPSAPQ 720 LSEERNGNGS GGCMEEGGSL LTVAFQILVN SLPTAKLTVE SVETVNNLIS CTVQKIKAAL 780 HCDSN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 97 | 102 | RKKKKR |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.1723 | 0.0 | flower | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in trichomes forming at the base of young leaves, in endodermal cell lines around emergent lateral roots and in the epidermal layer of the stamen filament. {ECO:0000269|PubMed:16778018}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00418 | DAP | Transfer from AT3G61150 | Download |
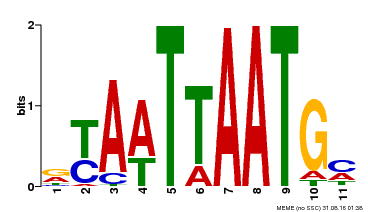
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009104292.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ182490 | 0.0 | DQ182490.1 Brassica napus baby boom interacting protein 2 mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009104292.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG1 | ||||
| Swissprot | Q9M2E8 | 0.0 | HDG1_ARATH; Homeobox-leucine zipper protein HDG1 | ||||
| TrEMBL | M4CGV6 | 0.0 | M4CGV6_BRARP; Uncharacterized protein | ||||
| STRING | Bra003439.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61150.1 | 0.0 | homeodomain GLABROUS 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



