 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009106897.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 682aa MW: 75428.6 Da PI: 7.2068 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.3 | 6.4e-19 | 42 | 95 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k +++t++q++eLe++F+ +++p++++r eL +kl+L+ +q+k+WFqNrR+++k
XP_009106897.1 42 KYRRHTSYQIQELESFFKVCPHPNEKQRLELGSKLSLESKQIKFWFQNRRTQMK 95
56789**********************************************999 PP
| |||||||
| 2 | START | 171.8 | 4.5e-54 | 207 | 425 | 3 | 205 |
HHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS...SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEECT CS
START 3 aeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv..dsgealrasgvvdmvlallveellddkeqWdetla....kaetleviss 87
a a++el+++a+++ p+W+k s +s + d ++++f+ sk + e +r++g+v ++ +lve+l+d++ +W e+++ a+t+evis+
XP_009106897.1 207 AAVAMDELIRLAEVDNPLWTKCSkserDSMKHDQYTSIFAGSKHpgFAAEGSRETGLVLINSSTLVETLMDTN-RWAEMFEcivaVASTVEVISN 300
56689******************9999999999******9988888888889*********************.********************* PP
T......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SS CS
START 88 g......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvdlkgr 175
g gal lm+ae+q++splvp R f+Ry++q+g+g w++vdvS d++++ + +s+ ++lpSg++i++ +ng skvtw+eh +++g+
XP_009106897.1 301 GsggsrnGALLLMQAEFQVMSPLVPiRQLKFLRYCKQHGDGLWAVVDVSYDVNRESQDLKSYGGLKRLPSGCIIQDIGNGCSKVTWIEHSEYEGS 395
*******************************************************99999*********************************** PP
XXHHHHHHHHHHHHHHHHHHHHHHTXXXXX CS
START 176 lphwllrslvksglaegaktwvatlqrqce 205
++h l+++l++s++ ga +w+atlqr ce
XP_009106897.1 396 HIHPLYQQLLGSSVGLGATKWLATLQRRCE 425
*****************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.88E-18 | 22 | 97 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-19 | 25 | 97 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 16.406 | 37 | 97 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.7E-15 | 39 | 101 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.3E-16 | 42 | 95 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.41E-15 | 42 | 97 | No hit | No description |
| PROSITE profile | PS50848 | 45.823 | 196 | 429 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.87E-31 | 200 | 427 | No hit | No description |
| CDD | cd08875 | 7.25E-105 | 201 | 425 | No hit | No description |
| SMART | SM00234 | 6.2E-43 | 205 | 426 | IPR002913 | START domain |
| Pfam | PF01852 | 5.2E-48 | 207 | 426 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 2.4E-4 | 210 | 409 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 2.64E-18 | 452 | 675 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 682 aa Download sequence Send to blast |
MNGDFDISKG DDEFESDSFE AMSGDDDDKH EQRPKKKKKM SKYRRHTSYQ IQELESFFKV 60 CPHPNEKQRL ELGSKLSLES KQIKFWFQNR RTQMKTQLER HENAILRQEN EKLRVENGIL 120 KEAMRSPPTC NNCGGAATPG NVSHDQQQLR MENAKLKYEL DKLCALANRF IGGSISLEQP 180 SNGGVGSQDL SLGHGFTRGT STFMDIAAVA MDELIRLAEV DNPLWTKCSK SERDSMKHDQ 240 YTSIFAGSKH PGFAAEGSRE TGLVLINSST LVETLMDTNR WAEMFECIVA VASTVEVISN 300 GSGGSRNGAL LLMQAEFQVM SPLVPIRQLK FLRYCKQHGD GLWAVVDVSY DVNRESQDLK 360 SYGGLKRLPS GCIIQDIGNG CSKVTWIEHS EYEGSHIHPL YQQLLGSSVG LGATKWLATL 420 QRRCESYTTL LSSQDQTGLP LAGTKSTLTL AQRMKRNFYS GITGSPIHKW EKLVAENVGQ 480 ETRILTRKSL QPSGVVLSAA TSMWLPVTQQ RLFEFLCDSK CRNQWDILCN GASMENMLLI 540 PQRQSEGRCV SLLQHAGKHQ NESSMLILQE TWSDASGALV VYAPVDVPST NMVMSGGDSA 600 NVALLPSGFS ISPDGSSSWS DQIDKNGGLV NHESKGCLLT VGFQILVNSV PTTKLNMESV 660 QTVNNLIACT IHKIKAALSI PA |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in cells around the base of leaf primordia, in the outermost 2 to 3 cell layers along the boundary between two leaf primordia. Expressed in lateral root primordia and tips, and in the epidermal boundaries of two cotyledons at heart-stage embryo. {ECO:0000269|PubMed:16778018}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds to the DNA sequence 5'-GCATTAAATGC-3'. {ECO:0000269|PubMed:16778018}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00559 | DAP | Transfer from AT5G52170 | Download |
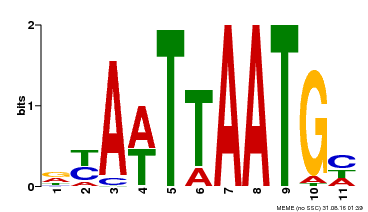
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009106897.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB025603 | 1e-103 | AB025603.1 Arabidopsis thaliana genomic DNA, chromosome 5, BAC clone:F17P19. | |||
| GenBank | CP002688 | 1e-103 | CP002688.1 Arabidopsis thaliana chromosome 5 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009106897.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG7 | ||||
| Swissprot | Q9LTK3 | 0.0 | HDG7_ARATH; Homeobox-leucine zipper protein HDG7 | ||||
| TrEMBL | M4EHN0 | 0.0 | M4EHN0_BRARP; Uncharacterized protein | ||||
| STRING | Bra028295.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G52170.1 | 0.0 | homeodomain GLABROUS 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



