 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009107887.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 909aa MW: 103421 Da PI: 7.7443 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 161.3 | 1.8e-50 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
+e +rwl+++ei+a+L n++ +++ ++ + pksg++++++rk++r+frkDG++wkkkkdgkt++E+he+LKvg+ e +++yYah+++nptf r
XP_009107887.1 30 EEaYTRWLRPNEIHALLSNHNYFTINVKPVHLPKSGTIVFFDRKMLRNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHVYYAHGDDNPTFVR 124
55589****************************************************************************************** PP
CG-1 97 rcywlLeeelekivlvhylevk 118
rcywlL++++e+ivlvhy+e++
XP_009107887.1 125 RCYWLLDKSQEHIVLVHYRETH 146
*******************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 75.622 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.2E-72 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.1E-44 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.26E-13 | 361 | 447 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.57E-13 | 535 | 657 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.9E-15 | 548 | 660 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 8.5E-7 | 548 | 627 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.71E-15 | 556 | 663 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 13.761 | 565 | 630 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.7E-6 | 598 | 627 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 598 | 630 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 7.181 | 747 | 773 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 260 | 762 | 784 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.3E-4 | 785 | 807 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.761 | 786 | 810 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 8.4E-5 | 787 | 807 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 11 | 861 | 883 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.334 | 862 | 891 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 909 aa Download sequence Send to blast |
MAGVDSGRLI GSEIHGFHTL QDLDIRTMLE EAYTRWLRPN EIHALLSNHN YFTINVKPVH 60 LPKSGTIVFF DRKMLRNFRK DGHNWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGDDNP 120 TFVRRCYWLL DKSQEHIVLV HYRETHEVQA APATPGNSYS SSTTDHVSAK PVTEDINSGV 180 RNACNTARSN SLVARNHEIS LHEINTLDWD ELLVETDDVL YFTEQLQTAA MGSAQQGNHH 240 AVYNGSTDIP SYLGLGDPVY QNNSPCGARE FSSQHLHCVV DPNQQTRDSS ATVADEQGDA 300 LLNNGYGSQE SFGKWVNNFI SDSPGSVDDP SLEAVYTPGQ ESSAPPAVVH SQSNIPEQVF 360 NITDVSPAWA YSTEKTKILV TGFFHDSFQH FGRSNLFCIC GELRVPAEFL QMGVYRCFLP 420 PQSPGIVNLY LSADGTKPIS QLFSFEHRSV PVIEKVVPQE DQLYKWEEFE FQVRLSHLLF 480 TSSSKISVFS SRISADNLLE AKKLASRTSH LLNSWAYLMK SIQANELPFD QARDPLFELT 540 LKNRLKEWLL EKVIENRNTK EYDSKGLGVI HLCAVLGYTW SILLFSWANI SLDFRDKHGW 600 TALHWAAYYG REKMVAALLS AGARPNLVTD PTKEYLGGCT AADIAQQKGY EGLAAFLAEK 660 CLVAQFRDMK MAGNISGNLE GVKAETSTNP GHSNEEEQSL KDTLAAYRTA AEAAARIQGA 720 FREHELKVRS KAVRFASKEE EAKNIIAAMK IQHAFRNYET RRKIAAAARI QYRFQTWKMR 780 REFLNMRKKA IKIQAVFRGF QVRRQYQKIT WSVGVLEKAI LRWRLKRRGF RGLQVSQPEE 840 KEGTEAVEDF YKTSQKQAED RLERSVVRVQ AMFRSKKAQQ DYRRMKLAHE EAQLEYDGMQ 900 ELNQMDIES |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.17287 | 0.0 | bud | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, pollen, top of sepals and siliques. {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
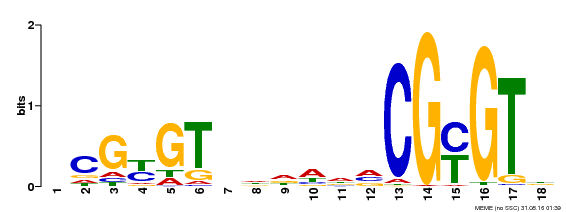
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009107887.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY128295 | 0.0 | AY128295.1 Arabidopsis thaliana AT4g16150/dl4115w mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009107887.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | M4FAC6 | 0.0 | M4FAC6_BRARP; Uncharacterized protein | ||||
| STRING | Bra038040.1-P | 0.0 | (Brassica rapa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



