 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009107928.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 491aa MW: 54894.5 Da PI: 6.29 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 21.1 | 8.1e-07 | 238 | 259 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
C++Cgk F+r nL+ H+r H
XP_009107928.1 238 FCTICGKGFKRDANLRMHMRGH 259
6*******************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 2.11E-5 | 235 | 262 | No hit | No description |
| PROSITE profile | PS50157 | 12.03 | 237 | 264 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0026 | 237 | 259 | IPR015880 | Zinc finger, C2H2-like |
| Pfam | PF12874 | 9.8E-4 | 238 | 259 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 6.8E-6 | 238 | 290 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 239 | 259 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 50 | 288 | 321 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010044 | Biological Process | response to aluminum ion | ||||
| GO:0010447 | Biological Process | response to acidic pH | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 491 aa Download sequence Send to blast |
MEPEDDLCQN NWGGSSSSSS SKRREQVCFT SQHKWEDASI LDYEMGMEEE PAFQENSNNN 60 NGGQVNVDFL QGVRAQAWDP RTMLSNLSFM EEKIHELQDL VHLMVARNGQ LQGRQEQLVA 120 QQQQLITTDL TSIIIQLIST AGSLLPSVKH HNMSTAPGPF TGSALFPYPR EANNLASQTL 180 NNNNNNTCEF DLPKPIVVEE EHEMKDEDDV EEGENLLPGS YEILQLEKEE ILAPHTHFCT 240 ICGKGFKRDA NLRMHMRGHG DEYKTPAALA KPNKEAVPGS EPMLIKRYSC PFPGCKRNKD 300 HKKFQPLKTI LCVKNHYKRT HCDKSFTRSR CHTKKFSVIA DLKTHEKHCG KNKWLCSCGT 360 TFSRKDKLFG HIALFQGHTP AIPLEETKPS AQKGSSACEN SNNNNTGMVG FNLGSATNAI 420 EEVAQPGFMD GKIRFEDSFS PLSFDTCNFG GFHEFPRPMF DDSESSFQML ISSACGFSPR 480 NGGESVSNTS L |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.605 | 6e-54 | bud| leaf| root| silique | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots (e.g. root tips and lateral roots), leaves, flowers (e.g. stigma, sepal, anther, and filament), stems, siliques and cotyledons. {ECO:0000269|PubMed:23935008}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Together with STOP2, plays a critical role in tolerance to major stress factors in acid soils such as proton H(+) and aluminum ion Al(3+). Required for the expression of genes in response to acidic stress (e.g. ALMT1 and MATE), and Al-activated citrate exudation. {ECO:0000269|PubMed:17535918, ECO:0000269|PubMed:18826429, ECO:0000269|PubMed:19321711, ECO:0000269|PubMed:23935008}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00196 | ampDAP | Transfer from AT1G34370 | Download |
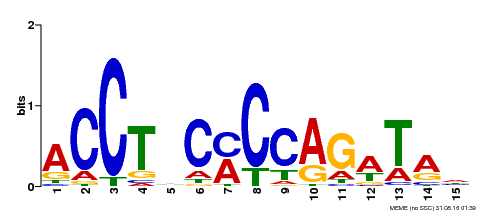
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009107928.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By shock H(+) and Al(3+) treatments. {ECO:0000269|PubMed:17535918}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC232513 | 0.0 | AC232513.1 Brassica rapa subsp. pekinensis cultivar Inbred line 'Chiifu' clone KBrB065E07, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009107928.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| Refseq | XP_018509453.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| Swissprot | Q9C8N5 | 0.0 | STOP1_ARATH; Protein SENSITIVE TO PROTON RHIZOTOXICITY 1 | ||||
| TrEMBL | M4F6P0 | 0.0 | M4F6P0_BRARP; Uncharacterized protein | ||||
| STRING | Bra036750.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6977 | 26 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34370.2 | 0.0 | C2H2 family protein | ||||



