 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009111469.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 790aa MW: 86496.6 Da PI: 6.2401 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.6 | 6.7e-21 | 124 | 179 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
XP_009111469.1 124 KKRYHRHTPQQIQELESMFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 179
688999***********************************************999 PP
| |||||||
| 2 | START | 203.4 | 9.2e-64 | 310 | 530 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS...SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv..dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevi 85
ela++a++elvk+a+++e++W ks +++n +e++++f+++k +ea+r+sg+v+ ++ lve+l+d+ +W+e+++ + +t++vi
XP_009111469.1 310 ELALTAMDELVKLAQSDEQLWIKSLdgerDELNHEEYMRTFSSTKPngLVTEASRTSGMVIINSLALVETLMDSD-RWTEMFPcivaRSTTTDVI 403
5899*************************************99999999**************************.******************* PP
CTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--..-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE- CS
START 86 ssg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe.sssvvRaellpSgiliepksnghskvtwvehvdl 172
s g ga+qlm+aelq+lsplvp R++ f+R+++q+ +g+w++vdvS+d ++++ s v+R +lpSg++++++sng+skvtwveh+++
XP_009111469.1 404 SGGmagtrnGAIQLMNAELQVLSPLVPvRNVNFLRFCKQHAEGVWAVVDVSIDTVRENSGgSTVVIR--RLPSGCVVQDMSNGYSKVTWVEHAEY 496
***********************************************************76666666..************************** PP
-SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 173 kgrlphwllrslvksglaegaktwvatlqrqcek 206
+++++h l+r+l++sgl +g ++wvatlqrqce+
XP_009111469.1 497 DENQIHHLYRPLLRSGLGFGSQRWVATLQRQCEC 530
********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.2E-20 | 111 | 181 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 7.3E-22 | 111 | 181 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.281 | 121 | 181 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.9E-18 | 122 | 185 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.49E-18 | 124 | 181 | No hit | No description |
| Pfam | PF00046 | 2.0E-18 | 124 | 179 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 156 | 179 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.623 | 301 | 533 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.63E-31 | 305 | 530 | No hit | No description |
| CDD | cd08875 | 1.60E-114 | 305 | 529 | No hit | No description |
| Pfam | PF01852 | 6.2E-57 | 310 | 530 | IPR002913 | START domain |
| SMART | SM00234 | 2.4E-39 | 310 | 530 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.21E-21 | 550 | 774 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 790 aa Download sequence Send to blast |
MNFGSLFDNT TGGVPTGLAY GNHTTASTII PGGTMSHIGA ASLFSPPINT KSVYSSSGLS 60 LALEQPERGT SMRNHNGAGG DIFDGSGKNR RSREEEHESR SGSDNVEGIS GEDQDADYNK 120 PPKKKRYHRH TPQQIQELES MFKECPHPDE KQRLELSKRL CLETRQVKFW FQNRRTQMKT 180 QLERHENALL RQENDKLRAE NMSIREAMRN PTCNICGGPA MLGEVSIEEH HLRIENARLK 240 DELDRVFNLT GKFLGHPQNN HTSSLELGVG TNNNGGNFAF PQDFNGGGGC LPPQLPAVVN 300 GVDQRSVLLE LALTAMDELV KLAQSDEQLW IKSLDGERDE LNHEEYMRTF SSTKPNGLVT 360 EASRTSGMVI INSLALVETL MDSDRWTEMF PCIVARSTTT DVISGGMAGT RNGAIQLMNA 420 ELQVLSPLVP VRNVNFLRFC KQHAEGVWAV VDVSIDTVRE NSGGSTVVIR RLPSGCVVQD 480 MSNGYSKVTW VEHAEYDENQ IHHLYRPLLR SGLGFGSQRW VATLQRQCEC LAILMSSSVT 540 SHDDTSISPG GRKSMLKLAQ RMTINFCSGI SAPSVHSWSK LTVGNVDPDV RIMTRKSVDD 600 PSEAPGIVLS AATSVWLPAS PQRLFYFLRN ERMRCEWDIL SNGGPMQEMA HIAKGQDQGN 660 SVSLLRSNPM NANQSSMLIL QETCIDASGA LVVYAPVDIP AMHVVMNGGD SSYVALLPSG 720 FAVLPDGGFN GGSGDGEQRP VGGGSLLTVA FQILVNNLPT AKLTVESVET VNNLISCTVQ 780 KIRTALQCEN |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves and floral buds. {ECO:0000269|PubMed:10402424}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
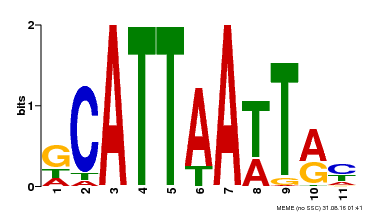
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009111469.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009111469.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2-like | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | M4F8E3 | 0.0 | M4F8E3_BRARP; Uncharacterized protein | ||||
| STRING | Bra037355.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



