 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009112123.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 999aa MW: 111818 Da PI: 6.0973 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 189 | 4.6e-59 | 19 | 136 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+e ++rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+e+n++fq
XP_009112123.1 19 LSEaQHRWLRPAEICEILRNYQKFHIASEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEDNENFQ 113
45559****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rrcyw+Le+el +iv+vhylevk
XP_009112123.1 114 RRCYWMLEQELMHIVFVHYLEVK 136
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 86.826 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 8.0E-87 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.6E-52 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.26E-12 | 401 | 485 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 2.64E-19 | 580 | 693 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 20.182 | 582 | 664 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.1E-18 | 582 | 694 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.9E-8 | 583 | 658 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 3.47E-16 | 588 | 691 | No hit | No description |
| PROSITE profile | PS50088 | 9.351 | 599 | 631 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1500 | 599 | 628 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.301 | 632 | 664 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 4.7E-4 | 632 | 661 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.32 | 809 | 831 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 9.56E-8 | 810 | 859 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.858 | 811 | 839 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0019 | 811 | 830 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 832 | 854 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.304 | 833 | 857 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.4E-4 | 835 | 854 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 999 aa Download sequence Send to blast |
MAYRGSFGFA PQLDIQQLLS EAQHRWLRPA EICEILRNYQ KFHIASEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDVLH CYYAHGEDNE NFQRRCYWML 120 EQELMHIVFV HYLEVKGNRI SSSGIKENNS NSLSGSTSVN IDSTANTSST LSPLCEDADS 180 GNRDGWIHGN RVKESDSQRL VGVPALDASF ENPLARYQNP PYNPLLTQTN PTNTGLMSVE 240 GHLRNPLQNQ VNWQIPVQDS LPLQKWPMDS HGTDLALHEN FGTFSSLIGS QNQQQPIGGG 300 SFQAPFTSVE AAYIPKFGPE DLLYEASANQ TLPLRKSLLK KEDSLKKVDS FSRWVSNELA 360 EMEDLQMQSS SGGIGWTSVE TAAAASSLSP SLSKDQRFTM IDFWPKWTQT DTEVEVMVIG 420 TFLLSPQEVT SYSWACMFGE VEVPAEILVD GVLCCHAPPH EVGQVPFYIT CSDRFSCSEV 480 REFDFLPGSA RKLNTVDIYG AYTNEASLHL RFENLLARMS SAQEHNVFED VGEKRRKISR 540 IMLLKDEKES FLTSTVEKDL TEVEAKERLI REEFEDKLYL WLIHKVTEEG KGPNILDEEG 600 QGVLHLAAAL GYDWAIKPIL AAGVSINFRD ANGWSALHWA AYSGREDTVA LLVSLGADSG 660 ALTDPSPELP LGKTASDLAY GNGHRGISGF LAESSLTSYL EKLTVDGKED ASTDSSRAKA 720 VQTVAERTAT PMSYGDVPET LSMKDSLTAV LNATQAADRL HQVFRMQSFQ RKQLSEIGDK 780 NEFGLSDELA VSFAAGKTKK AGGHSSGAAV HAAAVQIQKK YRGWKKRKEF LLIRQRIVKI 840 QAHVRGHQVR KQYRAIIWSV GLLEKIILRW RRKGSGLRGF KRDAVTKAPE PVCAAPAQED 900 DYDFLKEGRK QTEERLQKAL TRVKSMAQYP EARAQYRRLL TVVEGIRENE ASSSSAMNNN 960 NNNNNNSNTE EAANYNEEDD LIDIDSLLDD DTFMSLAFE |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.5064 | 0.0 | root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, old leaves, petals, sepals, top of carpels, stigmas, stamen filaments, anthers and siliques, but not in pollen. {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
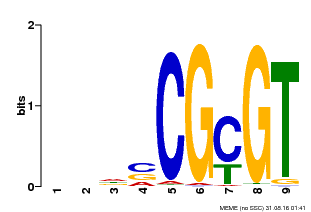
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009112123.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009112123.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A397XS62 | 0.0 | A0A397XS62_BRACM; Uncharacterized protein | ||||
| STRING | Bra037769.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



