 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009114751.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 560aa MW: 62126.2 Da PI: 4.3408 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 157.2 | 7e-49 | 13 | 139 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevls 94
+pGfrFhPtdeelv++yLk+k+ +++l++ ++i vd+yk++P++Lp + +k+++++w+fF++r++ky++ r+ r t++gyWkatgkd+ ++
XP_009114751.1 13 APGFRFHPTDEELVMYYLKRKICKTRLRV-SAIGVVDVYKLDPEELPgQsVLKSGDRQWFFFTPRNRKYPNAARSGRCTATGYWKATGKDRVIVY 106
69***************************.88**************96447788999*************************************9 PP
NAM 95 kkgelvglkktLvfykgrapkgektdWvmheyrl 128
+++ vglkktLvfy+grap+ge+tdWvmhey++
XP_009114751.1 107 -DSRSVGLKKTLVFYRGRAPNGERTDWVMHEYTM 139
.999****************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 52.455 | 12 | 162 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 2.09E-55 | 13 | 162 | IPR003441 | NAC domain |
| Pfam | PF02365 | 6.7E-26 | 14 | 138 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0070301 | Biological Process | cellular response to hydrogen peroxide | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 560 aa Download sequence Send to blast |
MADSWLKGGK LSAPGFRFHP TDEELVMYYL KRKICKTRLR VSAIGVVDVY KLDPEELPGQ 60 SVLKSGDRQW FFFTPRNRKY PNAARSGRCT ATGYWKATGK DRVIVYDSRS VGLKKTLVFY 120 RGRAPNGERT DWVMHEYTMD EDELGRCNTA KEYYALYKLF KKSGAGPKNG EDYGAPFQEE 180 EWVDDVNEVG DSNIPVNEES VTRRVDNATL FNPVNVQLDD VEEILNGIPY APGVPQTCIS 240 GLASIIPQLC LQVNSQEEEL PSTLVNNSSG DFLPNVQPYN MPSTFESTEA TSVPNDSGMG 300 PFVFEQEDYI EMDDLLTSEL GASSTEKPEQ LLDPGEFREF NEFDQLFHDV SMFLDMDPIL 360 QGTSADPSSL SNFANTSDQE DQSLYQQFQD QTPENKLNNI MDPNPNLSQF TDNLWFQDDY 420 QAVLFDQPHS IISGAFASPS SGVDVMPGST NLTTSVTAQD QEGENGGGGT SPFSSALCAF 480 MDSIPSTPAS ACEGPINRTF VRMSSFSRMR FGGTPATTTF VAKKRSRNRG FLVLSIVGAL 540 CAIFWVFIAT VRASGRPVFS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 6e-45 | 14 | 139 | 17 | 140 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 523 | 528 | KRSRNR |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.13695 | 0.0 | bud | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, rosette leaves, cauline leaves, shoot apex, stems and flowers. {ECO:0000269|PubMed:17158162}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP). Transcriptional activator that acts as positive regulator of AOX1A during mitochondrial dysfunction. Binds directly to AOX1A promoter. Mediates mitochondrial retrograde signaling. {ECO:0000269|PubMed:24045017}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00179 | DAP | Transfer from AT1G34190 | Download |
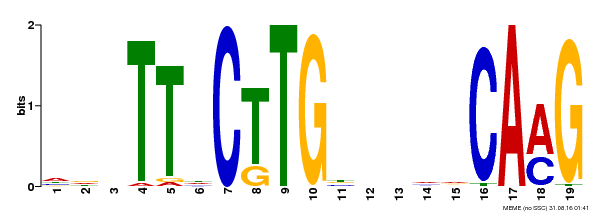
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009114751.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold and drought stresses. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009114751.1 | 0.0 | PREDICTED: NAC domain-containing protein 17-like isoform X1 | ||||
| Swissprot | Q9XIC5 | 0.0 | NAC17_ARATH; NAC domain-containing protein 17 | ||||
| TrEMBL | M4EGX4 | 0.0 | M4EGX4_BRARP; Uncharacterized protein | ||||
| STRING | Bra028039.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2812 | 26 | 67 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34190.1 | 0.0 | NAC domain containing protein 17 | ||||



