 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009122514.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1034aa MW: 116727 Da PI: 6.0767 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 184.4 | 1.3e-57 | 21 | 138 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+e ++rwl++ ei++iL n++k+++++e++trp sgsl+L++rk++ryfrkDG++w+kkkdgkt++E+hekLKvg+++vl+cyYah+e n++fq
XP_009122514.1 21 LSEaQHRWLRPAEICEILRNYHKFHIATESPTRPASGSLFLFDRKVLRYFRKDGHNWRKKKDGKTIKEAHEKLKVGSIDVLHCYYAHGEGNENFQ 115
45559****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rrcyw+Le el +iv+vhylevk
XP_009122514.1 116 RRCYWMLEVELMHIVFVHYLEVK 138
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 83.994 | 17 | 143 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 7.8E-84 | 20 | 138 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.6E-50 | 23 | 137 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.17E-13 | 416 | 502 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:2.60.40.10 | 9.0E-4 | 417 | 491 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF12796 | 9.6E-7 | 598 | 676 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-16 | 599 | 711 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 18.802 | 599 | 681 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.52E-17 | 600 | 710 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 8.15E-13 | 605 | 708 | No hit | No description |
| PROSITE profile | PS50088 | 11.22 | 649 | 681 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0029 | 649 | 678 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 6.37E-8 | 825 | 876 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 1.3 | 826 | 848 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.602 | 828 | 856 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0012 | 828 | 847 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0029 | 849 | 871 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.322 | 850 | 874 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.3E-4 | 852 | 871 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1034 aa Download sequence Send to blast |
MADRGSFGFI SPPRLDMEQL LSEAQHRWLR PAEICEILRN YHKFHIATES PTRPASGSLF 60 LFDRKVLRYF RKDGHNWRKK KDGKTIKEAH EKLKVGSIDV LHCYYAHGEG NENFQRRCYW 120 MLEVELMHIV FVHYLEVKGS RTSIGMKENN SNSLSGTASV NIDSAASPTS RLSSYCEDAD 180 SGDSHQSSSV LRASPEPQTG NRNGWTSAPG MRTVSQVHGN RVGETDSQRL FDVQTWDAVD 240 NLVTRYDQPC NNLLLEERTD KGGMLPAEHL RSPLQTQLNW QIPAQDDLPL PKWPGYLLPH 300 SGMTDDTDLA LLEQSAQDNF ESFSSLLDIE HLQSDGISPS DMESEYIPVK KSLLRHEDSL 360 KKVDSFSRWA SKELGEMEDL QMQSSRGDIA WTSVDCETAA AGVAFSPSLS EDQRFTILDY 420 WPKCAQTDAD VEVVVIGTFL LSPQEVTICS WSCMFGEVEV PAEILVDGVL CCHAPPHTAG 480 QVPFYVTCSN RFACSELREF DFRSGSTKKI DAAGIYGYST KEASLQMRFE KLLAHRDFVQ 540 EHQIFEDVVE KRRKISKIIL LNEEKENLFP RIYERHSTKQ EPKELVLRQQ FEDELYIWLI 600 HKVTEEGKGP NILDEGGQGV LHFVAALGYD WAIKPILAAG VNINFRDANG WSALHWAAFS 660 GREETVAVLV SLGADAGALT DPSPELPLGK TAADLAYGKE HRGISGFLAE SSLTSYLEKL 720 TMESKENSPA NSGGPKAVQT VSERTAAPMS SGDIPETLSL KDSLTAVRNA TQAADRLHQV 780 FRMQSFQRKQ LSGFDVDDDD EIGISNELAV SFAASKTKNP GQSEIFVHSA ATHIQKKYRG 840 WKKRKEFLLI RQRVVKIQAH VRGHQVRKQY KPIVWSVGLL EKIILRWRRK GTGLRGFKRN 900 AVPKTVEPEP QCPMVPKEDD YDFLEKGRKQ TEARLEKALT RVKSMVQYPE ARDQYRRLLT 960 VVEGFRENEA SSSLSVNNRE EPVNYEDDDL IDIDSLLNDD ILMSTSPXMT PFVYSFSFFR 1020 LSNFLHLFSV NIIS |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: During leaf developlement, expressed in young leaf primordia, hydathodes of young leaf tips, hydathodes of the lobes in maturating leaves and lamina within the minor veins in adult leaves. During flower development, expressed in developing stamens, germinating pollen grains, ovules and developing seeds. {ECO:0000269|PubMed:20383645}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, pollen and siliques. {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
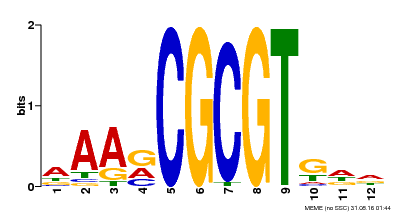
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009122514.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF491304 | 0.0 | AF491304.1 Brassica napus calmodulin-binding transcription activator (CAMTA) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009122514.2 | 0.0 | PREDICTED: calmodulin-binding transcription activator 1 | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | M4CYT5 | 0.0 | M4CYT5_BRARP; Uncharacterized protein | ||||
| STRING | Bra009382.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||



