 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009125041.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 269aa MW: 31221.8 Da PI: 6.5112 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 52.3 | 9.9e-17 | 75 | 127 | 4 | 56 |
-SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 4 RttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+ +++ eq+++Le+ Fe +++ e++ +LAk lg++ rq+ +WFqNrRa++k
XP_009125041.1 75 KKRLQLEQVKVLEKSFELGNKLEPERKIQLAKALGMQLRQIAIWFQNRRARWK 127
4457889*********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 119.6 | 1.6e-38 | 73 | 165 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekk+rl+ eqvk+LE+sFe +kLeperK +la++Lg+q rq+a+WFqnrRAR+kt+qlE+dy++Lk+++++lk++n++L +++++L +e+++
XP_009125041.1 73 EKKKRLQLEQVKVLEKSFELGNKLEPERKIQLAKALGMQLRQIAIWFQNRRARWKTRQLERDYDSLKKQFESLKSDNDSLLAHNKKLLAEVMS 165
69*************************************************************************************999875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.71E-18 | 69 | 131 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 1.5E-15 | 72 | 133 | IPR001356 | Homeobox domain |
| PROSITE profile | PS50071 | 15.742 | 73 | 129 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.10E-14 | 74 | 130 | No hit | No description |
| Pfam | PF00046 | 5.6E-14 | 75 | 127 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-17 | 77 | 137 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 6.1E-5 | 100 | 109 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 104 | 127 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 6.1E-5 | 109 | 125 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.0E-15 | 130 | 168 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 269 aa Download sequence Send to blast |
MALPQHAFMF QQRHEDNSHD QLPSCPPPHL FNGGGNYMMN RSMSLMNVQE DHHQSVDHEE 60 NLSDDGAHMM LGEKKKRLQL EQVKVLEKSF ELGNKLEPER KIQLAKALGM QLRQIAIWFQ 120 NRRARWKTRQ LERDYDSLKK QFESLKSDND SLLAHNKKLL AEVMSLKNND CNEGSIIKRE 180 AEASWSNNGS IENSSDINLE IPRETTTTPV STIKDLFPSS IRASTHHRQN HEMVQEESLC 240 NMFNGIEETT TDGYWAWSDP NHNNHRQFN |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed 3 day after germination (DAG) in first rosette leaf primordia around the veins in the apical part of the leaf and close to the midvein in the basal part of the leaf. At later stage, expressed in new forming veins, and veins and fascicular cambium of mature leaves. {ECO:0000269|PubMed:12644682}. | |||||
| Uniprot | TISSUE SPECIFICITY: Widely expressed. {ECO:0000269|PubMed:16055682}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00322 | DAP | Transfer from AT3G01220 | Download |
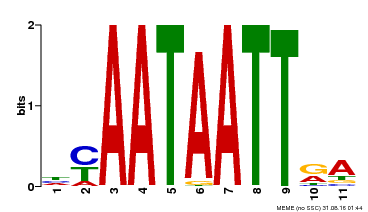
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009125041.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. {ECO:0000269|PubMed:12644682}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK228890 | 0.0 | AK228890.1 Arabidopsis thaliana mRNA for putative homeobox-leucine zipper protein, complete cds, clone: RAFL16-20-K05. | |||
| GenBank | AY087631 | 0.0 | AY087631.1 Arabidopsis thaliana clone 3722 mRNA, complete sequence. | |||
| GenBank | BT029221 | 0.0 | BT029221.1 Arabidopsis thaliana At3g01220 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009125041.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-20-like | ||||
| Swissprot | Q8LAT0 | 1e-167 | ATB20_ARATH; Homeobox-leucine zipper protein ATHB-20 | ||||
| TrEMBL | A0A398AUP0 | 0.0 | A0A398AUP0_BRACM; Uncharacterized protein | ||||
| TrEMBL | A0A3P5ZRK4 | 0.0 | A0A3P5ZRK4_BRACM; Uncharacterized protein (Fragment) | ||||
| STRING | scaffold_300109.1 | 1e-173 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3095 | 26 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01220.1 | 1e-169 | homeobox protein 20 | ||||



