 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009130506.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 310aa MW: 34227.1 Da PI: 4.5375 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.1 | 1.5e-18 | 73 | 126 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ q+++Le+ Fe ++++ e++ +LA++lgL+ rqV +WFqNrRa++k
XP_009130506.1 73 KKRRLSVVQVKALEKNFEIDNKLEPERKVKLAQELGLQPRQVAIWFQNRRARWK 126
55689999*********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 127.4 | 6e-41 | 72 | 164 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekkrrls qvk+LE++Fe ++kLeperKv+la+eLglqprqva+WFqnrRAR+ktkqlE+dy +Lk+++d+lk+++++L++++++L +e+ke
XP_009130506.1 72 EKKRRLSVVQVKALEKNFEIDNKLEPERKVKLAQELGLQPRQVAIWFQNRRARWKTKQLERDYGVLKSNFDSLKRSRDSLQRDNDSLLAEIKE 164
69**************************************************************************************99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.22E-19 | 66 | 130 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.248 | 68 | 128 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.4E-18 | 71 | 132 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.99E-18 | 73 | 129 | No hit | No description |
| Pfam | PF00046 | 9.6E-16 | 73 | 126 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-19 | 75 | 135 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 7.7E-6 | 99 | 108 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 103 | 126 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 7.7E-6 | 108 | 124 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 3.0E-17 | 128 | 169 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 310 aa Download sequence Send to blast |
MKRSRGSSDS LSGFLPICHS TADKQLSPRP TATGFLYPGG AGDYSQMFDG LEEDGSLEDI 60 GVGHASSTAA AEKKRRLSVV QVKALEKNFE IDNKLEPERK VKLAQELGLQ PRQVAIWFQN 120 RRARWKTKQL ERDYGVLKSN FDSLKRSRDS LQRDNDSLLA EIKELRAKLD VEGTCGNNGN 180 AVTEETGVVK PVETVAFQTV IANNEVLELS QCPPLPGEAP ASELAYEMFS IFPRTESFRE 240 DPADSSDSSA VLNEEYSPTA AAATAVEMST MGCFGQFVKM EEHEDLFSGE EACKLFADNE 300 QWFCSGQWSS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 120 | 128 | RRARWKTKQ |
| 2 | 141 | 146 | DSLKRS |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.2958 | 0.0 | bud| leaf| root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Localized primarily to the hypocotyl of germinating seedlings. {ECO:0000269|PubMed:12678559}. | |||||
| Uniprot | TISSUE SPECIFICITY: Widely expressed. {ECO:0000269|PubMed:16055682}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
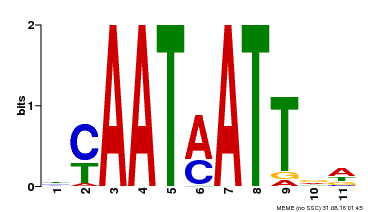
| UniProt | Probable transcription factor that acts as a positive regulator of ABA-responsiveness, mediating the inhibitory effect of ABA on growth during seedling establishment. Binds to the DNA sequence 5'-CAATNATTG-3'. {ECO:0000269|PubMed:12678559}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00587 | DAP | Transfer from AT5G65310 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009130506.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by abscisic acid (ABA) and by salt stress. {ECO:0000269|PubMed:12678559, ECO:0000269|PubMed:16055682}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF325054 | 0.0 | AF325054.2 Arabidopsis thaliana AT5g65310 (AT5g65310/MNA5_4) mRNA, complete cds. | |||
| GenBank | AY074293 | 0.0 | AY074293.1 Arabidopsis thaliana putative homeobox-leucine zipper protein ATHB-5 (At5g65310) mRNA, complete cds. | |||
| GenBank | AY091340 | 0.0 | AY091340.1 Arabidopsis thaliana putative homeobox-leucine zipper protein ATHB-5 (HD-zip protein ATHB-5) (At5g65310) mRNA, complete cds. | |||
| GenBank | X67033 | 0.0 | X67033.1 A.thaliana mRNA Athb-5. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009130506.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-5-like | ||||
| Refseq | XP_013678412.1 | 0.0 | homeobox-leucine zipper protein ATHB-5-like | ||||
| Swissprot | P46667 | 1e-165 | ATHB5_ARATH; Homeobox-leucine zipper protein ATHB-5 | ||||
| TrEMBL | A0A398ANU2 | 0.0 | A0A398ANU2_BRACM; Uncharacterized protein | ||||
| TrEMBL | M4EST6 | 0.0 | M4EST6_BRARP; Uncharacterized protein | ||||
| STRING | Bra031866.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM12986 | 15 | 26 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65310.1 | 1e-167 | homeobox protein 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



