 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009138054.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 571aa MW: 64235.9 Da PI: 6.4365 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 81.5 | 9.2e-26 | 192 | 245 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr+ Wt eLH++F+ av++L G ekA Pk+ilelm+v+ Lt+e+v+SHLQk+R+
XP_009138054.1 192 KPRVLWTRELHNKFLAAVDHL-GVEKAQPKKILELMNVDKLTRENVASHLQKFRS 245
79*******************.********************************7 PP
| |||||||
| 2 | Response_reg | 88.6 | 1.7e-29 | 19 | 127 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaG 93
vl vdD+++ +++l+++l + +y +v+++d+++ alell+e++ +Dl++ D++mp+mdG++ll+ e++lp+i+++ah++++++ + +k G
XP_009138054.1 19 VLAVDDDQTCLRILETLLHRCQY-HVTTTDSAQTALELLRENKnkFDLVISDVDMPDMDGFKLLELVGL-EMDLPVIMLSAHSDPKYVMKGVKHG 111
799********************.***************888888*******************87754.558********************** PP
ESEEEESS--HHHHHH CS
Response_reg 94 akdflsKpfdpeelvk 109
a d+l Kp+ +eel +
XP_009138054.1 112 ACDYLLKPVRIEELKN 127
*************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF52172 | 8.68E-37 | 16 | 137 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 2.6E-43 | 16 | 169 | No hit | No description |
| SMART | SM00448 | 2.1E-34 | 17 | 129 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 43.87 | 18 | 133 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 2.4E-26 | 19 | 127 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 1.07E-29 | 20 | 132 | No hit | No description |
| PROSITE profile | PS51294 | 14.564 | 189 | 248 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-27 | 189 | 250 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.78E-19 | 190 | 249 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.8E-24 | 192 | 245 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.5E-6 | 196 | 243 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0031537 | Biological Process | regulation of anthocyanin metabolic process | ||||
| GO:0048367 | Biological Process | shoot system development | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0080036 | Biological Process | regulation of cytokinin-activated signaling pathway | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 571 aa Download sequence Send to blast |
MTLEQDFEAV DQFPVGMRVL AVDDDQTCLR ILETLLHRCQ YHVTTTDSAQ TALELLRENK 60 NKFDLVISDV DMPDMDGFKL LELVGLEMDL PVIMLSAHSD PKYVMKGVKH GACDYLLKPV 120 RIEELKNIWQ HVVRKSKFKK MKSIVINDDH SQGNSDQNGV KANRKRKDQF EEVEEEDEER 180 GNENDDPTAQ KKPRVLWTRE LHNKFLAAVD HLGVEKAQPK KILELMNVDK LTRENVASHL 240 QKFRSALKKI TNEANQQANM AAIDSHFMQM SALKGLGGFH NQRQIPLGSG QFHGGAATMR 300 HYPLGRLNSF GGVFPHVSSS LPRNHNDGGY VLQGMPIPPL DDLNNKAFPS FTSQQSSLMV 360 APNNQLVLQG HQQSSYPSLN PGLSPHFEIN KRLDDWSNAL LSTNIPQSGV HSKPDALEWN 420 HFCNSDAAQA GFIDPLQMKQ QPANNLGPMT DAQLLRSSNP IEGLFVGQQK LENGSMPSNA 480 GSLDDIVNSM MPKEQSQAEL FEGDWGLDGI IAHSEHAYEN LFYVFYVLGQ HEKECFLQTQ 540 SFSIIFGSGL FVCSLFVKKL QALVYKFPRR F |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-31 | 188 | 251 | 1 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.14192 | 0.0 | flower| root| seed | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Detected in the whole plant. Predominantly expressed in roots and leaves. {ECO:0000269|PubMed:15173562}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins. {ECO:0000269|PubMed:11574878}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00072 | SELEX | Transfer from AT4G31920 | Download |
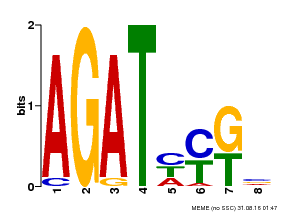
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009138054.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353533 | 0.0 | AK353533.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-50-C17. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009138054.1 | 0.0 | PREDICTED: LOW QUALITY PROTEIN: two-component response regulator ARR10 | ||||
| Swissprot | O49397 | 0.0 | ARR10_ARATH; Two-component response regulator ARR10 | ||||
| TrEMBL | M4E5B8 | 0.0 | M4E5B8_BRARP; Two-component response regulator | ||||
| STRING | Bra023972.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1981 | 28 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31920.1 | 0.0 | response regulator 10 | ||||



