 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009138294.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 270aa MW: 29680.8 Da PI: 6.7714 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 54.2 | 3.6e-17 | 139 | 188 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
gy+GVr+++ +g+++AeIrdp+ r +r++lg f tae+Aa+a+++a+ ++g
XP_009138294.1 139 GYRGVRQRP-WGKFAAEIRDPK----RaTRVWLGPFETAEDAARAYDRAAIGFRG 188
8********.**********83....57*********************987776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00380 | 1.9E-35 | 139 | 202 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.393 | 139 | 196 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 9.3E-11 | 139 | 187 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.19E-21 | 139 | 197 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 3.93E-20 | 140 | 196 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 1.2E-30 | 140 | 196 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.2E-10 | 140 | 151 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.2E-10 | 162 | 178 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 270 aa Download sequence Send to blast |
MHYPNNTRPE FVGAPASERD STRNRYQQKE QLSPEQELSV IVSALQHVLS GEKETTPYQG 60 FSSSSTVISA GMHSDTCQVC RIDGCLGCNY FFAPNQRIEK KQQVEVEEGV TSKSSGREST 120 VVAAAATGKI RKRKNKKNGY RGVRQRPWGK FAAEIRDPKR ATRVWLGPFE TAEDAARAYD 180 RAAIGFRGPR AKLNFPFVDY TSATTSSSVP AEDTRTNASA SVSAVDSVEA EQWRGGEGGD 240 CDMEEYLKMV MDFGNGDSSD SGNTIADMFQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 4e-24 | 136 | 197 | 2 | 64 | ATERF1 |
| 3gcc_A | 4e-24 | 136 | 197 | 2 | 64 | ATERF1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.28999 | 3e-91 | leaf| root | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00469 | DAP | Transfer from AT4G34410 | Download |
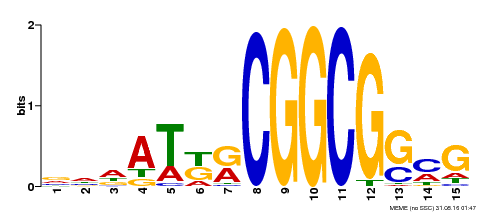
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009138294.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009138294.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor ERF109-like | ||||
| Swissprot | Q9SZ06 | 1e-106 | EF109_ARATH; Ethylene-responsive transcription factor ERF109 | ||||
| TrEMBL | M4DMC3 | 0.0 | M4DMC3_BRARP; Uncharacterized protein | ||||
| STRING | Bra017656.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5469 | 24 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G34410.1 | 8e-92 | redox responsive transcription factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



