 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009140343.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1031aa MW: 115187 Da PI: 5.3387 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 181.6 | 9.2e-57 | 20 | 136 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
+e ++rwl++ ei++iL+n++k+++++e++t+p sgs++L++rk++ryfrkDG++w+kk+dgktv+E+he+LK g+v+vl+cyYah+++n++fqr
XP_009140343.1 20 SEaRNRWLRPPEICEILQNYQKFQISTEPPTTPASGSVFLFDRKVLRYFRKDGHNWRKKRDGKTVKEAHERLKAGSVDVLHCYYAHGQDNENFQR 114
4449******************************************************************************************* PP
CG-1 97 rcywlLeeelekivlvhylevk 118
r+yw+L+eel++iv+vhylevk
XP_009140343.1 115 RSYWMLQEELSHIVFVHYLEVK 136
*******************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.969 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.0E-84 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 9.6E-50 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.12E-15 | 457 | 539 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 4.05E-11 | 618 | 748 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 3.1E-15 | 619 | 752 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 17.184 | 622 | 748 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.71E-15 | 634 | 751 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.11 | 689 | 721 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 8.5E-9 | 799 | 902 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.92 | 851 | 873 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.029 | 853 | 881 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0041 | 854 | 872 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0011 | 874 | 896 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.468 | 875 | 899 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.2E-4 | 877 | 896 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1031 aa Download sequence Send to blast |
MAEARRFGLN NELDVGQILS EARNRWLRPP EICEILQNYQ KFQISTEPPT TPASGSVFLF 60 DRKVLRYFRK DGHNWRKKRD GKTVKEAHER LKAGSVDVLH CYYAHGQDNE NFQRRSYWML 120 QEELSHIVFV HYLEVKGSRV STSYNRMQRT EDSTRSSQET GEVYTSERNG YASGSINQYD 180 HSNNQSQATD SASVNGVHTP ELEDAQSAYN QQGSPILYSH QALQQPPATS FDPYYQMSLT 240 PRDSYQKEIH TISSSTMVEK GRTINGPVVT NSIKNKKSID SQTWEEILGN CGSGGEGLPM 300 QPHSEHEGLD QMLQSYSFTM QDFASLQESI VKSQNQELNS GLTSDRSLWL QGQAVDIEPN 360 ALSNLASSEK APYLSTMKQH LLDGALGEEG LKKMDSFNRW MSKELGELGD VGVTADANES 420 FTHSSSTAYW EEVESEDVSN GGYVMSPSLS KEQLFSIIDF APNWTYVGCE VKVLVSGKFL 480 KMAESGEWCC MFGQTEVPAD IIANGILECV APMHEAGRVP FYVTCSNRLA CSEVREFEYK 540 VLESQGFDRE TYDSSTGCNS IESLEARFVK LLCSKSDCTN SSLPGGNDSD LSQVSEKISL 600 LLFENDDQLD QMLMNEISQE NMKNNLLQEA LKESLHSWLL QKIAEGGKGP NVLDEGGQGV 660 LHFAAALGYN WALEPTIVAG VSVDFRDVNG WTALHWAAFF GRELIIGSLI ALGASPGTLT 720 DPNPDFPSGS TPSDLAYANG YKGIAGYLSE YALRTHVSLL SLNEKNAETS LGGAVEAAPS 780 PSSSALTDSL TAVRNASQAA ARIHQVFRAQ SFQKKQMKEF GDRKLGMSEE RALSMLAPKT 840 HKQGRGHSDD SVQAAAIRIQ NKFRGYKGRK DYLITRQRII KIQAHVRGYQ VRKNYRKIIW 900 SVGILEKVIL RWRRKGAGLR GFKSDALVTK MQDGTEKEED DDFFKQGRKQ TEERLEKALA 960 RVKSMVQYPE ARDQYRRLLN VVNDIQESKV EKALANSEEA TCFDDDLIDI EALLGDDDTL 1020 MMPMSSTLWN A |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, carpels, and siliques, but not in stigmas or other parts of the flower. {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
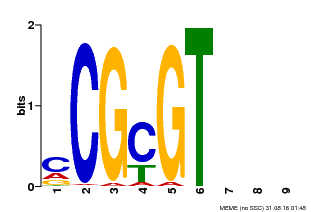
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009140343.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009140343.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 3-like | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | M4EN78 | 0.0 | M4EN78_BRARP; Uncharacterized protein | ||||
| STRING | Bra030248.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8601 | 25 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||



