 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009142669.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 355aa MW: 38189.1 Da PI: 9.962 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 72 | 9e-23 | 237 | 297 | 1 | 61 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklks 61
e+elkrerrkq+NRe+ArrsR+RK+ae+eeL++kv++L+aeN+aL++el++l++++ +l+
XP_009142669.1 237 ERELKRERRKQSNRESARRSRLRKQAETEELARKVEALTAENMALRSELNQLNEKSNNLRG 297
89*****************************************************999976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 6.5E-31 | 1 | 96 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 1.3E-14 | 130 | 232 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 4.7E-17 | 236 | 297 | No hit | No description |
| Pfam | PF00170 | 5.1E-21 | 237 | 299 | IPR004827 | Basic-leucine zipper domain |
| SMART | SM00338 | 3.0E-21 | 237 | 301 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.346 | 239 | 302 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 6.25E-11 | 240 | 296 | No hit | No description |
| CDD | cd14702 | 3.25E-22 | 242 | 293 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 244 | 259 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 355 aa Download sequence Send to blast |
MGKSEEPKVT KSDNKPSSPP ADQTNVHVYP DWAAMQAYYG PRVAMPPYYN SAMAAASGHP 60 PPPYMWNPQH MMSPYGTPYA AVYPHGGGVY AHPGFPMGSQ PHGQKGATPL TTPGTPLNID 120 TPSKSTGNTE NGLMKKLKEF DGLAMSLGNG NNGDEGKRSR NSSETDGSSD GSDGNTTGAD 180 EPKLKRSREG TPAKDEKKHL VQSSSFRSVS QSSGDNNCVK PSVQGGGGAI VSAAGNEREL 240 KRERRKQSNR ESARRSRLRK QAETEELARK VEALTAENMA LRSELNQLNE KSNNLRGANA 300 TLLDKLKSSE PEKRVKSSGN GDDKNKKQGD NETNSTSKLH QLLDTKPRAD GVAAR |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 253 | 259 | RRSRLRK |
| 2 | 253 | 260 | RRSRLRKQ |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.30061 | 0.0 | root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Present only in dark grown leaves and roots. {ECO:0000269|PubMed:8672884}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the G-box motif (5'-CCACGTGG-3') of the rbcS-1A gene promoter. G-box and G-box-like motifs are cis-acting elements defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control. {ECO:0000269|PubMed:8672884}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00318 | DAP | Transfer from AT2G46270 | Download |
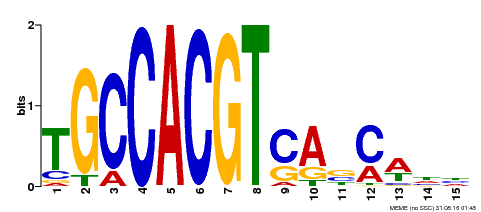
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009142669.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BNU27108 | 0.0 | U27108.1 Brassica napus transcription factor (BnGBF1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009142669.1 | 0.0 | PREDICTED: G-box-binding factor 3 isoform X2 | ||||
| Swissprot | P42776 | 0.0 | GBF3_ARATH; G-box-binding factor 3 | ||||
| TrEMBL | M4CK16 | 0.0 | M4CK16_BRARP; Uncharacterized protein | ||||
| STRING | Bra004550.1-P | 0.0 | (Brassica rapa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46270.2 | 1e-170 | G-box binding factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



