 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009145103.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 326aa MW: 36960.6 Da PI: 5.9686 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 99 | 3.3e-31 | 193 | 248 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++WtpeLH+ Fv+a++qLGG+ +AtPk+i+e+m+ +gLt+++vkSHLQkYRl+
XP_009145103.1 193 KQRRCWTPELHRLFVDALQQLGGPGVATPKQIREHMQEEGLTNDEVKSHLQKYRLH 248
79****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.565 | 190 | 250 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.15E-18 | 191 | 251 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-27 | 192 | 251 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.5E-26 | 193 | 248 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.9E-9 | 195 | 246 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MGSLGDELSL GQRRHSSIPT TICRSKIMNV AVEQGCKLEE HVQKLEEEKR KIQGSELELP 60 LCLQMLNDAI SYLKKETETD TQPLLKDFIS LSKPIREEHD EELFKEKKFQ LWRENDRISN 120 ARFSDTLEIE RDEEKSSNSL YMLLSPGMTT PKVETGLGLG LTSSSMVKGR RKLVASSSVP 180 QPPPYLQQQS LRKQRRCWTP ELHRLFVDAL QQLGGPGVAT PKQIREHMQE EGLTNDEVKS 240 HLQKYRLHIR KSDSNLEKQS VVVLGFNLWN SSQEVEESGE GGETSKRNNS QSDSPQGPLQ 300 LPCTTTTTCG DSSMEDAEDA KSERRY |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate signaling in roots. {ECO:0000250|UniProtKB:Q9FX67}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00193 | DAP | Transfer from AT1G49560 | Download |
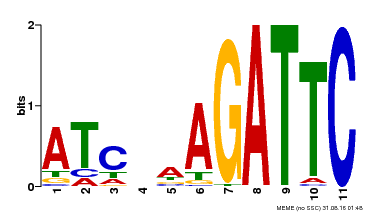
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009145103.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK221631 | 1e-113 | AK221631.1 Arabidopsis thaliana mRNA for hypothetical protein, complete cds, clone: RAFL07-96-G04. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009145103.1 | 0.0 | PREDICTED: myb family transcription factor EFM-like | ||||
| Swissprot | Q9FX84 | 1e-122 | HHO6_ARATH; Transcription factor HHO6 | ||||
| TrEMBL | M4FHT4 | 0.0 | M4FHT4_BRARP; Uncharacterized protein | ||||
| STRING | Bra040662.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13927 | 18 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49560.1 | 1e-102 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



