 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009146834.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 455aa MW: 51180.3 Da PI: 5.8445 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 168.5 | 2.3e-52 | 30 | 157 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevl 93
l+pGfrFhPtdeelv++yLk+kv gk++++ ++i +vdiyk+ePwdL +++k++++ewyf+s dkky +g r nrat++gyWkatgkd+e+
XP_009146834.1 30 LAPGFRFHPTDEELVSYYLKRKVMGKPVRF-DAIGDVDIYKHEPWDLAvfSRLKTRDQEWYFYSALDKKYGNGARMNRATNKGYWKATGKDREIR 123
579************************999.99**************8544899999************************************** PP
NAM 94 skkgelvglkktLvfykgrapkgektdWvmheyrl 128
+ + +l g+kktLvf++grap+g +t+Wvmheyrl
XP_009146834.1 124 R-DIQLLGMKKTLVFHSGRAPDGLRTNWVMHEYRL 157
9.999****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.7E-59 | 23 | 182 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.244 | 30 | 181 | IPR003441 | NAC domain |
| Pfam | PF02365 | 8.6E-27 | 32 | 157 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 455 aa Download sequence Send to blast |
MGRESVAVVS SPPTATATAR GTATVAATSL APGFRFHPTD EELVSYYLKR KVMGKPVRFD 60 AIGDVDIYKH EPWDLAVFSR LKTRDQEWYF YSALDKKYGN GARMNRATNK GYWKATGKDR 120 EIRRDIQLLG MKKTLVFHSG RAPDGLRTNW VMHEYRLVDY ETESNGNLVQ DAYVLCRVFH 180 KNNIGPPSGN RYAPFLEEEW ADEGVALIPG VDVRVRAEPL PVANGNNQMD QEIQSASKDL 240 ININEPPRET TPMDIEVNHQ NHHENAPKPQ ENNNNNHYDE AEEALKLEQA EEDERPPPAC 300 VLNKEAPLPL LQYKRRRQNE PNNSSRTTQD HCSSTITTVD NTTPTLISSS AAAATNTAIS 360 ALLEFSLMGI SAKKENTPQP PNKEAYPPAP LPSTEEKLND LQKEVHQMSV ERETFKLEMM 420 SAEAMISILQ SRIDALRQEN DELRKNNATK GQAVL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 7e-47 | 22 | 183 | 12 | 170 | NAC domain-containing protein 19 |
| 3swm_B | 7e-47 | 22 | 183 | 12 | 170 | NAC domain-containing protein 19 |
| 3swm_C | 7e-47 | 22 | 183 | 12 | 170 | NAC domain-containing protein 19 |
| 3swm_D | 7e-47 | 22 | 183 | 12 | 170 | NAC domain-containing protein 19 |
| 3swp_A | 7e-47 | 22 | 183 | 12 | 170 | NAC domain-containing protein 19 |
| 3swp_B | 7e-47 | 22 | 183 | 12 | 170 | NAC domain-containing protein 19 |
| 3swp_C | 7e-47 | 22 | 183 | 12 | 170 | NAC domain-containing protein 19 |
| 3swp_D | 7e-47 | 22 | 183 | 12 | 170 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Bra.12284 | 0.0 | bud | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Mostly expressed in floral organs, and, at low levels, in other organs. {ECO:0000269|PubMed:25578968}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
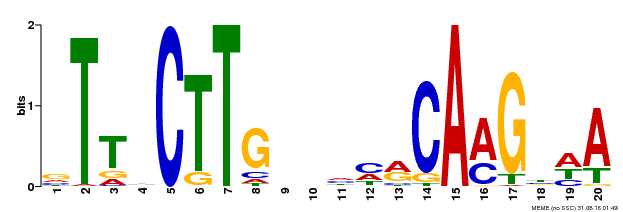
| UniProt | Transcriptional repressor that binds to the motif 5'-(C/T)A(C/A)G-3' in the promoter of target genes (PubMed:25578968). Binds also to the 5'-CTTGNNNNNCAAG-3' consensus sequence in chromatin (PubMed:26617990). Can bind to the mitochondrial dysfunction motif (MDM) present in the upstream regions of mitochondrial dysfunction stimulon (MDS) genes involved in mitochondrial retrograde regulation (MRR) (PubMed:24045019). Together with NAC051/NAC052 and JMJ14, regulates gene expression and flowering time by associating with the histone demethylase JMJ14, probably by the promotion of RNA-mediated gene silencing (PubMed:25578968, PubMed:26617990). {ECO:0000269|PubMed:24045019, ECO:0000269|PubMed:25578968, ECO:0000269|PubMed:26617990}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00612 | ChIP-seq | Transfer from AT3G10490 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009146834.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009146834.1 | 0.0 | PREDICTED: NAC domain-containing protein 13-like isoform X1 | ||||
| Swissprot | Q9SQX9 | 0.0 | NAC50_ARATH; NAC domain containing protein 50 | ||||
| TrEMBL | M4EM24 | 0.0 | M4EM24_BRARP; Uncharacterized protein | ||||
| STRING | Bra029844.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1780 | 28 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G10480.1 | 0.0 | NAC domain containing protein 50 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



