 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009149819.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1385aa MW: 154692 Da PI: 7.1782 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.2 | 0.0012 | 1268 | 1293 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++sFs+ +L H r+ +
XP_009149819.1 1268 YQCDmeGCTMSFSSEKQLSLHKRNiC 1293
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 15.1 | 6.8e-05 | 1293 | 1315 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H+r+H
XP_009149819.1 1293 CPvkGCGKNFFSHKYLVQHQRVH 1315
9999*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 4.1E-17 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.284 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 1.7E-15 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 1.7E-54 | 198 | 367 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 34.513 | 201 | 367 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.24E-27 | 213 | 384 | No hit | No description |
| Pfam | PF02373 | 2.4E-37 | 231 | 350 | IPR003347 | JmjC domain |
| Gene3D | G3DSA:3.30.160.60 | 0.001 | 1267 | 1290 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 9.4 | 1268 | 1290 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.01 | 1291 | 1315 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.609 | 1291 | 1320 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.1E-7 | 1292 | 1319 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1293 | 1315 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.91E-11 | 1307 | 1349 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.4E-10 | 1320 | 1344 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0015 | 1321 | 1345 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.801 | 1321 | 1350 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1323 | 1345 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.95E-8 | 1339 | 1373 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 7.0E-10 | 1345 | 1374 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 1.7 | 1351 | 1377 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.492 | 1351 | 1382 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1353 | 1377 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1385 aa Download sequence Send to blast |
MAVSEQSQDV FPWLKSLPVA PEFRPTLAEF QDPMAYICKI EEEASRYGIC KIVPPVPPSS 60 KKTAINNLNR SLAARARARA RDGGKPDYDG GPTFTTRQQQ IGFCPRRQRP VQRPVWQSGE 120 RYTFDEFEFK AKNFEKSYLK KCGKKGSVSP LEVETLYWRA SVDKPFSVEY ANDMPGSAFV 180 PLSLAAARRR ESGGDCGTVG ETAWNMRAMA RAEGSLLKFM KEEIPGVTSP MVYIAMMFSW 240 FAWHVEDHDL HSLNYLHMGA GKTWYGVPKD AAMAFEEVVR VHGYGEELNP LVTFSTLGEK 300 TTVMSPEVFV RAGIPCCRLV QNPGDFVITF PGAYHSGFSH GFNFGEASNI ATPQWLRMAK 360 DAAIRRASIN YPPMVSHLQL LYDYALALGS RVPDSIHNKP RSSRLKDKKK SEGEKLTNEL 420 FVQNIIHNNE LLHSLGKGSP IALLPQSSSD VSVCSDVRIG SHLGANQGQT TLLIKSEDLS 480 SDSVMVSLSN GVKEKFTSLC ERNRDHLASK EDETQGTSTD VERRKNNGAV GLSDQRLFSC 540 VTCGVLSFDC VAIIQPKEAA ARYLMSADCS FLNDWTVASG SGNLGQDVVM PPSENTGKQD 600 VGDLYNAPVQ TPYHSTTKTV DQRTSSSSLT KESGALGLLA SAYGDSSDSE EEDHKGLDNP 660 VSDEVACVLE ASSFVTDGND EAGNGLSSAL NSQGLTCEKG KEVDVSHANL SKGGNASSVE 720 ITLPFIPRSD DDFSRLHVFC LEHAAEVEQR LHPIGGINIM LLCHPDYPRI EAEGKVVAKE 780 LGVNHEWNYT EFKNVTREDE ETIQAALANV EAKAGNSDWA VKLGINLSHS AILSRSPLYS 840 KQMPYNSVIY NVFGRTSPAT PQVSGIRSSR QRKYVAGKWC GKVWMSHQVH PFLLEEDFEG 900 EETERSHLRA ALDEDVTVHG NDSRDATTMF GRKYSRKRKA RGKAAPRKKL TSFKREAGVS 960 DDTSEDHSYK QQWRAYGDEE ESYFETGNEV SGDSSNQMSD QQQLKGVESD DDEVSERSLG 1020 QEYTVRREYA SSESSKENGF QVYREDQPMY GDNDMYRHPR GIPRSKRTKV FRDLVSYDSE 1080 DQQRERVFTS DAQTSRMDDE YDSEENSLEE QDFCSSGKRQ TRSIAKRKVK TKIVQSLGDT 1140 GGRTLLQSGS RKKMKELDSY MEGPSTRLRV RTPKPSRGSS ATKPKKTGKK GRNVSFSEVA 1200 SEEEVEENEE ESEEASTRIR VRTLKPPRRS LETKPKKTGK KGSNVSSLSG VASEEEVEED 1260 EQEECLPYQC DMEGCTMSFS SEKQLSLHKR NICPVKGCGK NFFSHKYLVQ HQRVHSDERP 1320 LKCPWKGCKM TFKWAWSRTE HIRVHTGERP YVCAEPGCSQ TFRFVSDFSR HKRKTGHSAK 1380 KIKKK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 7e-74 | 1265 | 1385 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a58_A | 7e-74 | 1265 | 1385 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a59_A | 7e-74 | 1265 | 1385 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6ip0_A | 2e-70 | 9 | 390 | 4 | 355 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 2e-70 | 9 | 390 | 4 | 355 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 936 | 948 | KRKARGKAAPRKK |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Highly expressed in the shoot apical meristem and primary and secondary root tips, and lower expression in cotyledons, leaves and root axis along vascular tissues. Detected in inflorescences, stems and siliques. {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18713399}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
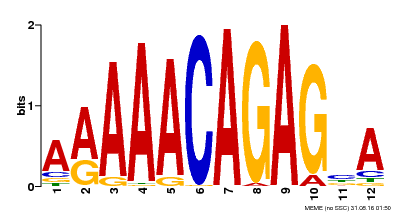
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_009149819.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009149819.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 isoform X1 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A397YY33 | 0.0 | A0A397YY33_BRACM; Uncharacterized protein | ||||
| TrEMBL | M4DNH3 | 0.0 | M4DNH3_BRARP; Uncharacterized protein | ||||
| STRING | Bra018060.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



