 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009587298.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 717aa MW: 79142.9 Da PI: 6.5942 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61.1 | 1.7e-19 | 29 | 84 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q+++Le+ F+++++p++++r +L++ lgL rq+k+WFqNrR+++k
XP_009587298.1 29 KKRYHRHTANQIQRLEAIFKECPHPDEKTRLQLSRDLGLAPRQIKFWFQNRRTQMK 84
678899***********************************************998 PP
| |||||||
| 2 | START | 180.5 | 9.6e-57 | 225 | 450 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEE CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla....kaetl 82
+a a++el+++++ +ep+W+ks+ + +n + + ++f+++++ ++ea+r+sgvv+m+ lv ++d + +W+e ++ ka tl
XP_009587298.1 225 IAGNAMEELIRLVQTNEPLWMKSAidgkDVLNFQNYDRIFPRANShlknhnVRIEASRDSGVVIMNGLALVDMFMDAN-KWQEFFPtivsKARTL 318
67899*********************99999*********999999**9999**************************.**************** PP
EEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-E CS
START 83 evissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehv 170
evissg ++lqlm+ elq+lsplvp R +f+R+++q ++ +w+ivdvS d +q++ ss + ++++lpSg+li++++ng+skvtwvehv
XP_009587298.1 319 EVISSGvmgsrtSTLQLMYEELQVLSPLVPtRQLYFLRFCQQIEQSSWAIVDVSYDITQENLYSSTSCKVHRLPSGCLIQDMPNGYSKVTWVEHV 413
************************************************************977******************************** PP
E--SSXX.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 171 dlkgrlp.hwllrslvksglaegaktwvatlqrqcek 206
+++++ + h l r l++sgla+ga +wv tlqr cek
XP_009587298.1 414 EVEEKGIiHRLCRDLIHSGLAFGAERWVGTLQRVCEK 450
***99988***************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.17E-19 | 11 | 86 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 8.4E-22 | 14 | 88 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.475 | 26 | 86 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.0E-18 | 27 | 90 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 7.35E-18 | 28 | 87 | No hit | No description |
| Pfam | PF00046 | 3.7E-17 | 29 | 84 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 61 | 84 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 48.911 | 215 | 453 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 4.12E-34 | 216 | 452 | No hit | No description |
| CDD | cd08875 | 1.68E-115 | 219 | 449 | No hit | No description |
| SMART | SM00234 | 2.1E-48 | 224 | 450 | IPR002913 | START domain |
| Pfam | PF01852 | 9.8E-49 | 225 | 450 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.14E-22 | 471 | 707 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 717 aa Download sequence Send to blast |
MEYGSGGGGG GGGTSSGGDD PRDAAERRKK RYHRHTANQI QRLEAIFKEC PHPDEKTRLQ 60 LSRDLGLAPR QIKFWFQNRR TQMKAQHERA DNCALRAEND KIRCENIAIR EALKNVICPS 120 CPPVTEDSYF DEQKLRIENM QLKEELDKVS SIAAKYIGRP ISQLPPVQPI HLSSLDLSMS 180 SFVGHGPNSL DLDLDLLPGS SLTIPSLAFC SLNLSDMDKS LMADIAGNAM EELIRLVQTN 240 EPLWMKSAID GKDVLNFQNY DRIFPRANSH LKNHNVRIEA SRDSGVVIMN GLALVDMFMD 300 ANKWQEFFPT IVSKARTLEV ISSGVMGSRT STLQLMYEEL QVLSPLVPTR QLYFLRFCQQ 360 IEQSSWAIVD VSYDITQENL YSSTSCKVHR LPSGCLIQDM PNGYSKVTWV EHVEVEEKGI 420 IHRLCRDLIH SGLAFGAERW VGTLQRVCEK YACLMVNSNP THDLGGVIPS PEGKRSMMKL 480 AKRMVSNFCA SINPSNGHQW NTISGLNEFE VRATLQKSTD PGQPNGVIIS AATTIWLPVP 540 PQNVFNFFRD ERNRPQWDVL SNQNPVQEVA HIANGSHPGN CISVLRAYNT SQNNMLILQE 600 SCIDSSGALV VYSPVDVPAI NIAMSGEDPT YIPLLPSGFT ISPDGHQLDQ DAASCSSSSN 660 ASTIGGRSGG SLITVVFQIL VSSLPSAKMS PESVNTVNNL IGSTVHQIKA ALNCSTS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which acts as positive regulator of drought stress tolerance. Can transactivate CIPK3, NCED3 and ERECTA (PubMed:18451323). Transactivates several cell-wall-loosening protein genes by directly binding to HD motifs in their promoters. These target genes play important roles in coordinating cell-wall extensibility with root development and growth (PubMed:24821957). Transactivates CYP74A/AOS, AOC3, OPR3 and 4CLL5/OPCL1 genes by directly binding to HD motifs in their promoters. These target genes are involved in jasmonate (JA) biosynthesis, and JA signaling affects root architecture by activating auxin signaling, which promotes lateral root formation (PubMed:25752924). Acts as negative regulator of trichome branching (PubMed:16778018, PubMed:24824485). Required for the establishment of giant cell identity on the abaxial side of sepals (PubMed:23095885). May regulate cell differentiation and proliferation during root and shoot meristem development (PubMed:25564655). {ECO:0000269|PubMed:16778018, ECO:0000269|PubMed:18451323, ECO:0000269|PubMed:23095885, ECO:0000269|PubMed:24821957, ECO:0000269|PubMed:24824485, ECO:0000269|PubMed:25564655, ECO:0000269|PubMed:25752924}. | |||||
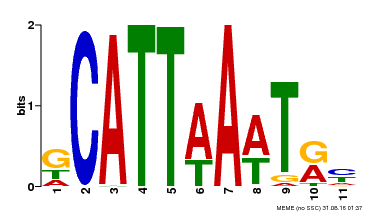
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975442 | 1e-160 | HG975442.1 Solanum pennellii chromosome ch03, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009587298.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG11-like | ||||
| Swissprot | Q9FX31 | 0.0 | HDG11_ARATH; Homeobox-leucine zipper protein HDG11 | ||||
| TrEMBL | A0A1S3XNG8 | 0.0 | A0A1S3XNG8_TOBAC; homeobox-leucine zipper protein HDG11-like isoform X2 | ||||
| STRING | XP_009587298.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2490 | 23 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



