 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009600619.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1055aa MW: 118444 Da PI: 5.7339 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 181.2 | 1.2e-56 | 20 | 136 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
+e ++rwl++ ei++iL n++k+++t e+++rp sgs++L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+ee+ +fqr
XP_009600619.1 20 SEvQHRWLRPAEICEILRNYKKFHITPEAPHRPVSGSVFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEEDDNFQR 114
5559******************************************************************************************* PP
CG-1 97 rcywlLeeelekivlvhylevk 118
r+yw+Le++l +iv+vhylevk
XP_009600619.1 115 RSYWMLEQDLMHIVFVHYLEVK 136
*******************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 81.799 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.2E-79 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 7.5E-50 | 22 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 7.1E-6 | 467 | 555 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 2.18E-17 | 469 | 555 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 8.7E-7 | 469 | 554 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 8.86E-18 | 651 | 763 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.9E-17 | 652 | 765 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 18.961 | 652 | 773 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.71E-15 | 665 | 761 | No hit | No description |
| PROSITE profile | PS50088 | 8.816 | 669 | 701 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 3400 | 669 | 698 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0036 | 702 | 731 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.484 | 702 | 734 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 900 | 741 | 770 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 5.31E-7 | 876 | 927 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 5 | 876 | 898 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.968 | 877 | 906 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0049 | 878 | 897 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 9.4E-4 | 899 | 921 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.56 | 900 | 924 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 7.6E-5 | 902 | 921 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1055 aa Download sequence Send to blast |
MADCGSDPSG FRLDITQILS EVQHRWLRPA EICEILRNYK KFHITPEAPH RPVSGSVFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDVLH CYYAHGEEDD NFQRRSYWML 120 EQDLMHIVFV HYLEVKGNKA NMGCVRSIKS AHSNYLNDCS LSDSFPRGHK KLASANADST 180 SVASTLTSAH EEAESDSHQA CSRFQSYPER ASGMDRNLVE NRDTIYSSYG SPQSSVEYTS 240 LPGIDVGEKC GLGNFASGPQ RTIDLGSQEP VSQHCSNGEI VCQDDFKNNL SVKGNWQYSF 300 GDSASQFHGQ IVNQDLIADS SYDLVNSFHN KNLSSDLYTG RGQSYLYPDE QEEQLTQLNI 360 QYLNSLVEVQ GDFNQENSMD MLGLGDYSTI KHPHLNSVKM EEGLKKVDSF SRWVVKELED 420 VEELHMQPTN RISWNVIDTE DDGSCLPTQL HVDSDSLNPS LSQEQVFSII DFSPNWAYSN 480 LETKVLITGR FLKSEGELIE CKWSCMFGEV EVPAEVLADG VLRCHAPPHK PGVLPFYVTC 540 SNRLACSEVR EFEYRLGAYQ EIGAANVSAT EMHLLERIES LMSLGPVSSC HSSDSMEAAK 600 EKHSTVNKII CMMEEENQQM IERASDYDTS QCGVKEDLFL ERKLKQNFYA WLVRQVTDDG 660 RGRTAIDDEG QGVLHLAAAL GYDWALKPIL ASGVSVDFRD MNGWTALHWA AFYGREKTVV 720 GLVSLGASPG ALTDPSAEFP LGRTPADLAS ANGHKGISGF LAESSLTTHL SKLTVTDATE 780 ELASEVSGAK VGETVTERVA VTTTGDDVPD VLSLKDSLAA IRNATQAAAR IHQIFRVQSF 840 QRKQIIERSD NELSSDENAL SIVASRACKL GQNNGIAHAA ATQIQKKFRG WNKRKEFLLI 900 RQKIVKIQAH VRGHQVRKKY KPIIWSVGIL EKVILRWRRK RSGLRGFRSE VVINKPSIQD 960 DSLPEDDYDF LKEGRKQTEV RMQKALARVK SMTQYPEGRA QYRRLLTAAE GLREVKQDGS 1020 TCIQESSEDT SYPEEELFDV ENLLDDDTFM SIAFE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
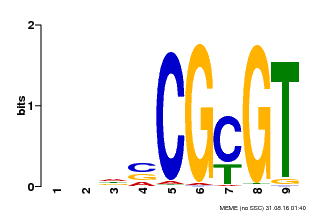
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN558810 | 0.0 | JN558810.1 Solanum lycopersicum calmodulin-binding transcription factor SR1L mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009600619.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2-like isoform X3 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A1S4BAF3 | 0.0 | A0A1S4BAF3_TOBAC; calmodulin-binding transcription activator 2-like isoform X1 | ||||
| STRING | XP_009600617.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



